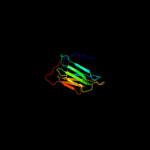
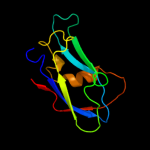
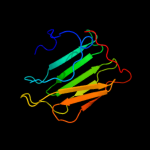
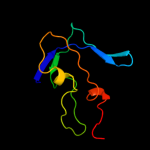
1 d1q56a_
46.5
11
Fold: Concanavalin A-like lectins/glucanasesSuperfamily: Concanavalin A-like lectins/glucanasesFamily: Laminin G-like module2 d1wmxa_
40.4
20
Fold: Galactose-binding domain-likeSuperfamily: Galactose-binding domain-likeFamily: Family 30 carbohydrate binding module, CBM30 (PKD repeat)3 c3pveA_
27.6
12
PDB header: transcriptionChain: A: PDB Molecule: agrin, agrn protein;PDBTitle: crystal structure of the g2 domain of agrin from mus musculus
4 d1uzva_
27.0
27
Fold: Calcium-mediated lectinSuperfamily: Calcium-mediated lectinFamily: Calcium-mediated lectin5 c2kfwA_
25.8
13
PDB header: isomeraseChain: A: PDB Molecule: fkbp-type peptidyl-prolyl cis-trans isomerasePDBTitle: solution structure of full-length slyd from e.coli
6 d1pz7a_
22.9
10
Fold: Concanavalin A-like lectins/glucanasesSuperfamily: Concanavalin A-like lectins/glucanasesFamily: Laminin G-like module7 d2chha1
22.0
20
Fold: Calcium-mediated lectinSuperfamily: Calcium-mediated lectinFamily: Calcium-mediated lectin8 d1ylxa1
20.0
21
Fold: N domain of copper amine oxidase-likeSuperfamily: GK1464-likeFamily: GK1464-like9 c2boiA_
17.9
10
PDB header: lectinChain: A: PDB Molecule: cv-iil lectin;PDBTitle: 1.1a structure of chromobacterium violaceum lectin cv2l in2 complex with alpha-methyl-fucoside
10 d1wmxb_
16.6
23
Fold: Galactose-binding domain-likeSuperfamily: Galactose-binding domain-likeFamily: Family 30 carbohydrate binding module, CBM30 (PKD repeat)11 d2d3na1
16.4
16
Fold: Glycosyl hydrolase domainSuperfamily: Glycosyl hydrolase domainFamily: alpha-Amylases, C-terminal beta-sheet domain12 c2r0fA_
16.0
11
PDB header: sugar binding proteinChain: A: PDB Molecule: cgl3 lectin;PDBTitle: ligand free structure of fungal lectin cgl3
13 d2bosa_
15.7
24
Fold: OB-foldSuperfamily: Bacterial enterotoxinsFamily: Bacterial AB5 toxins, B-subunits14 c2khgA_
15.1
42
PDB header: antimicrobial proteinChain: A: PDB Molecule: plnj;PDBTitle: plantaricin j in tfe
15 c2khfA_
14.9
42
PDB header: antimicrobial proteinChain: A: PDB Molecule: plnj;PDBTitle: plantaricin j in dpc-micelles
16 c1w9iA_
14.0
15
PDB header: myosinChain: A: PDB Molecule: myosin ii heavy chain;PDBTitle: myosin ii dictyostelium discoideum motor domain s456y bound2 with mgadp-befx
17 c2eg5C_
13.7
20
PDB header: transferaseChain: C: PDB Molecule: xanthosine methyltransferase;PDBTitle: the structure of xanthosine methyltransferase
18 c1qu0A_
13.5
9
PDB header: metal binding proteinChain: A: PDB Molecule: laminin alpha2 chain;PDBTitle: crystal structure of the fifth laminin g-like module of the2 mouse laminin alpha2 chain
19 c1qu0D_
13.5
9
PDB header: metal binding proteinChain: D: PDB Molecule: laminin alpha2 chain;PDBTitle: crystal structure of the fifth laminin g-like module of the2 mouse laminin alpha2 chain
20 d1ud2a1
13.4
11
Fold: Glycosyl hydrolase domainSuperfamily: Glycosyl hydrolase domainFamily: alpha-Amylases, C-terminal beta-sheet domain21 d1ulea_
not modelled
13.4
15
Fold: Concanavalin A-like lectins/glucanasesSuperfamily: Concanavalin A-like lectins/glucanasesFamily: Galectin (animal S-lectin)22 d2anua1
not modelled
13.3
16
Fold: 7-stranded beta/alpha barrelSuperfamily: PHP domain-likeFamily: PHP domain23 c2anuA_
not modelled
13.3
16
PDB header: metal binding proteinChain: A: PDB Molecule: hypothetical protein tm0559;PDBTitle: crystal structure of predicted metal-dependent phosphoesterase (php2 family) (tm0559) from thermotoga maritima at 2.40 a resolution
24 d2aw2a1
not modelled
13.3
14
Fold: Immunoglobulin-like beta-sandwichSuperfamily: ImmunoglobulinFamily: V set domains (antibody variable domain-like)25 d1hvxa1
not modelled
12.9
22
Fold: Glycosyl hydrolase domainSuperfamily: Glycosyl hydrolase domainFamily: alpha-Amylases, C-terminal beta-sheet domain26 c2wjsA_
not modelled
12.8
13
PDB header: cell adhesionChain: A: PDB Molecule: laminin subunit alpha-2;PDBTitle: crystal structure of the lg1-3 region of the laminin alpha22 chain
27 c2fvnA_
not modelled
12.8
28
PDB header: cell adhesionChain: A: PDB Molecule: protein afad;PDBTitle: the fibrillar tip complex of the afa/dr adhesins from2 pathogen e. coli displays synergistic binding to 5 1 and v3 3 integrins
28 c3hqxA_
not modelled
12.6
12
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: upf0345 protein aciad0356;PDBTitle: crystal structure of protein of unknown function (duf1255,pf06865)2 from acinetobacter sp. adp1
29 d1fyxa_
not modelled
12.6
25
Fold: Flavodoxin-likeSuperfamily: Toll/Interleukin receptor TIR domainFamily: Toll/Interleukin receptor TIR domain30 c3eo6B_
not modelled
12.5
14
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: protein of unknown function (duf1255);PDBTitle: crystal structure of protein of unknown function (duf1255)2 (afe_2634) from acidithiobacillus ferrooxidans ncib8455 at3 0.97 a resolution
31 c2zw7A_
not modelled
12.5
25
PDB header: transferaseChain: A: PDB Molecule: bleomycin acetyltransferase;PDBTitle: crystal structure of bleomycin n-acetyltransferase complexed2 with bleomycin a2 and coenzyme a
32 d2gjpa1
not modelled
12.3
16
Fold: Glycosyl hydrolase domainSuperfamily: Glycosyl hydrolase domainFamily: alpha-Amylases, C-terminal beta-sheet domain33 d1r4pb_
not modelled
11.7
33
Fold: OB-foldSuperfamily: Bacterial enterotoxinsFamily: Bacterial AB5 toxins, B-subunits34 d1c4qa_
not modelled
11.7
33
Fold: OB-foldSuperfamily: Bacterial enterotoxinsFamily: Bacterial AB5 toxins, B-subunits35 d2nn8a1
not modelled
11.7
26
Fold: Concanavalin A-like lectins/glucanasesSuperfamily: Concanavalin A-like lectins/glucanasesFamily: Galectin (animal S-lectin)36 d1dyka2
not modelled
11.5
10
Fold: Concanavalin A-like lectins/glucanasesSuperfamily: Concanavalin A-like lectins/glucanasesFamily: Laminin G-like module37 c3nv4A_
not modelled
11.3
20
PDB header: sugar binding proteinChain: A: PDB Molecule: galectin 9 short isoform variant;PDBTitle: crystal structure of human galectin-9 c-terminal crd in complex with2 sialyllactose
38 c3efyB_
not modelled
10.4
12
PDB header: cell cycleChain: B: PDB Molecule: cif (cell cycle inhibiting factor);PDBTitle: structure of the cyclomodulin cif from pathogenic2 escherichia coli
39 d2axwa1
not modelled
9.4
19
Fold: Common fold of diphtheria toxin/transcription factors/cytochrome fSuperfamily: Bacterial adhesinsFamily: Pilus subunits40 c3eitB_
not modelled
9.3
12
PDB header: unknown functionChain: B: PDB Molecule: putative atp/gtp binding protein;PDBTitle: the 2.6 angstrom crystal structure of chbp, the cif homologue from2 burkholderia pseudomallei
41 d1fyva_
not modelled
8.9
25
Fold: Flavodoxin-likeSuperfamily: Toll/Interleukin receptor TIR domainFamily: Toll/Interleukin receptor TIR domain42 d2oyza1
not modelled
8.9
25
Fold: Double-stranded beta-helixSuperfamily: RmlC-like cupinsFamily: VPA0057-like43 c3b5iB_
not modelled
8.8
22
PDB header: transferaseChain: B: PDB Molecule: s-adenosyl-l-methionine:salicylic acid carboxylPDBTitle: crystal structure of indole-3-acetic acid methyltransferase
44 c2eg9B_
not modelled
8.7
13
PDB header: hydrolaseChain: B: PDB Molecule: adp-ribosyl cyclase 1;PDBTitle: crystal structure of the truncated extracellular domain of2 mouse cd38
45 d1wdja_
not modelled
8.7
15
Fold: Restriction endonuclease-likeSuperfamily: Restriction endonuclease-likeFamily: Hypothetical protein TT1808 (TTHA1514)46 d1e43a1
not modelled
8.0
11
Fold: Glycosyl hydrolase domainSuperfamily: Glycosyl hydrolase domainFamily: alpha-Amylases, C-terminal beta-sheet domain47 c1qu0C_
not modelled
7.6
9
PDB header: metal binding proteinChain: C: PDB Molecule: laminin alpha2 chain;PDBTitle: crystal structure of the fifth laminin g-like module of the2 mouse laminin alpha2 chain
48 d2ftxb1
not modelled
7.4
13
Fold: Kinetochore globular domain-likeSuperfamily: Kinetochore globular domainFamily: Spc24-like49 c3b9cB_
not modelled
7.4
16
PDB header: unknown functionChain: B: PDB Molecule: hspc159;PDBTitle: crystal structure of human grp crd
50 c2fv4B_
not modelled
7.3
13
PDB header: structural protein, protein bindingChain: B: PDB Molecule: hypothetical 24.6 kda protein in ilv2-ade17PDBTitle: nmr solution structure of the yeast kinetochore spc24/spc252 globular domain
51 d2gala_
not modelled
6.5
25
Fold: Concanavalin A-like lectins/glucanasesSuperfamily: Concanavalin A-like lectins/glucanasesFamily: Galectin (animal S-lectin)52 c2yroA_
not modelled
6.5
20
PDB header: sugar binding proteinChain: A: PDB Molecule: galectin-8;PDBTitle: solution structure of the c-terminal gal-bind lectin2 protein from human galectin-8
53 d2uz9a1
not modelled
6.4
36
Fold: Composite domain of metallo-dependent hydrolasesSuperfamily: Composite domain of metallo-dependent hydrolasesFamily: SAH/MTA deaminase-like54 c3sh5A_
not modelled
6.1
11
PDB header: metal binding proteinChain: A: PDB Molecule: lg3 peptide;PDBTitle: calcium-bound laminin g like domain 3 from human perlecan
55 c2c5dA_
not modelled
6.1
9
PDB header: signaling protein/receptorChain: A: PDB Molecule: growth-arrest-specific protein 6 precursor;PDBTitle: structure of a minimal gas6-axl complex
56 d1wdjb_
not modelled
6.0
16
Fold: Restriction endonuclease-likeSuperfamily: Restriction endonuclease-likeFamily: Hypothetical protein TT1808 (TTHA1514)57 d2zgwa2
not modelled
5.9
36
Fold: Class II aaRS and biotin synthetasesSuperfamily: Class II aaRS and biotin synthetasesFamily: Biotin holoenzyme synthetase58 c2eayB_
not modelled
5.9
45
PDB header: ligaseChain: B: PDB Molecule: biotin [acetyl-coa-carboxylase] ligase;PDBTitle: crystal structure of biotin protein ligase from aquifex2 aeolicus
59 c3dtdI_
not modelled
5.8
13
PDB header: structural genomics, unknown functionChain: I: PDB Molecule: invasion-associated protein b;PDBTitle: crystal structure of invasion associated protein b from bartonella2 henselae
60 c2xr4A_
not modelled
5.8
30
PDB header: sugar binding proteinChain: A: PDB Molecule: lectin;PDBTitle: c-terminal domain of bc2l-c lectin from burkholderia cenocepacia
61 d2h3ka1
not modelled
5.6
26
Fold: Immunoglobulin-like beta-sandwichSuperfamily: NEAT domain-likeFamily: NEAT domain62 d1xaua_
not modelled
5.5
15
Fold: Immunoglobulin-like beta-sandwichSuperfamily: ImmunoglobulinFamily: I set domains