| 1 |
|
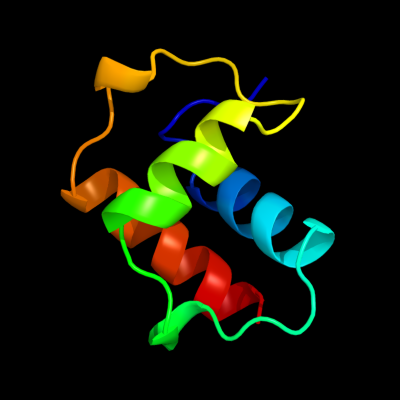
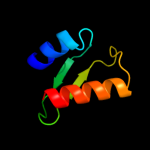
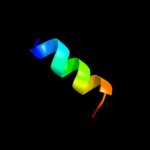
PDB 1uj8 chain A domain 1
Region: 2 - 65
Aligned: 64
Modelled: 64
Confidence: 100.0%
Identity: 100%
Fold: Another 3-helical bundle
Superfamily: IscX-like
Family: IscX-like
Phyre2
| 2 |
|
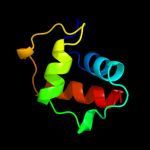
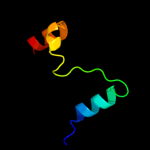
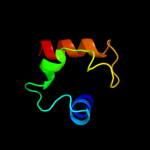
PDB 2kzv chain A
Region: 5 - 43
Aligned: 39
Modelled: 39
Confidence: 82.1%
Identity: 21%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: solution nmr structure of cv_0373(175-257) protein from2 chromobacterium violaceum, northeast structural genomics consortium3 target cvr118a
Phyre2
| 3 |
|
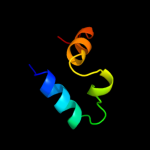
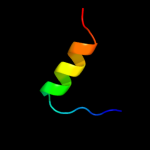
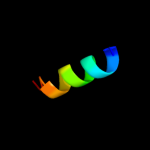
PDB 2kpm chain A
Region: 5 - 42
Aligned: 38
Modelled: 38
Confidence: 39.7%
Identity: 18%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: solution nmr structure of uncharacterized protein from gene2 locus ne0665 of nitrosomonas europaea. northeast structural3 genomics target ner103a
Phyre2
| 4 |
|
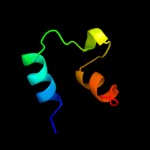
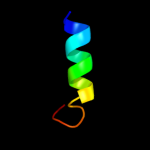
PDB 3gr1 chain A
Region: 8 - 62
Aligned: 55
Modelled: 55
Confidence: 20.4%
Identity: 16%
PDB header:membrane protein
Chain: A: PDB Molecule:protein prgh;
PDBTitle: periplamic domain of the t3ss inner membrane protein prgh2 from s.typhimurium (fragment 170-392)
Phyre2
| 5 |
|
PDB 1gwc chain A domain 2
Region: 7 - 24
Aligned: 18
Modelled: 18
Confidence: 15.6%
Identity: 17%
Fold: Thioredoxin fold
Superfamily: Thioredoxin-like
Family: Glutathione S-transferase (GST), N-terminal domain
Phyre2
| 6 |
|
PDB 3ktb chain D
Region: 20 - 60
Aligned: 41
Modelled: 41
Confidence: 12.7%
Identity: 17%
PDB header:transcription regulator
Chain: D: PDB Molecule:arsenical resistance operon trans-acting repressor;
PDBTitle: crystal structure of arsenical resistance operon trans-acting2 repressor from bacteroides vulgatus atcc 8482
Phyre2
| 7 |
|
PDB 3mg9 chain A
Region: 5 - 57
Aligned: 53
Modelled: 53
Confidence: 9.3%
Identity: 19%
PDB header:transferase/antibiotic
Chain: A: PDB Molecule:teg12;
PDBTitle: teg 12 binary structure complexed with the teicoplanin aglycone
Phyre2
| 8 |
|
PDB 1mzb chain A
Region: 4 - 27
Aligned: 24
Modelled: 24
Confidence: 8.0%
Identity: 25%
Fold: DNA/RNA-binding 3-helical bundle
Superfamily: "Winged helix" DNA-binding domain
Family: FUR-like
Phyre2
| 9 |
|
PDB 1n2a chain A domain 2
Region: 2 - 21
Aligned: 20
Modelled: 20
Confidence: 7.8%
Identity: 35%
Fold: Thioredoxin fold
Superfamily: Thioredoxin-like
Family: Glutathione S-transferase (GST), N-terminal domain
Phyre2
| 10 |
|
PDB 1gnw chain A domain 2
Region: 7 - 24
Aligned: 18
Modelled: 18
Confidence: 7.8%
Identity: 22%
Fold: Thioredoxin fold
Superfamily: Thioredoxin-like
Family: Glutathione S-transferase (GST), N-terminal domain
Phyre2
| 11 |
|
PDB 1e6b chain A domain 2
Region: 7 - 21
Aligned: 15
Modelled: 15
Confidence: 7.7%
Identity: 40%
Fold: Thioredoxin fold
Superfamily: Thioredoxin-like
Family: Glutathione S-transferase (GST), N-terminal domain
Phyre2
| 12 |
|
PDB 1fw1 chain A domain 2
Region: 7 - 21
Aligned: 15
Modelled: 15
Confidence: 7.0%
Identity: 33%
Fold: Thioredoxin fold
Superfamily: Thioredoxin-like
Family: Glutathione S-transferase (GST), N-terminal domain
Phyre2
| 13 |
|
PDB 2o03 chain A
Region: 7 - 27
Aligned: 21
Modelled: 21
Confidence: 6.6%
Identity: 19%
PDB header:gene regulation
Chain: A: PDB Molecule:probable zinc uptake regulation protein furb;
PDBTitle: crystal structure of furb from m. tuberculosis- a zinc uptake2 regulator
Phyre2
| 14 |
|
PDB 12as chain A
Region: 12 - 26
Aligned: 15
Modelled: 15
Confidence: 6.3%
Identity: 47%
Fold: Class II aaRS and biotin synthetases
Superfamily: Class II aaRS and biotin synthetases
Family: Class II aminoacyl-tRNA synthetase (aaRS)-like, catalytic domain
Phyre2
| 15 |
|
PDB 2xig chain A
Region: 8 - 27
Aligned: 20
Modelled: 20
Confidence: 6.2%
Identity: 15%
PDB header:transcription
Chain: A: PDB Molecule:ferric uptake regulation protein;
PDBTitle: the structure of the helicobacter pylori ferric uptake2 regulator fur reveals three functional metal binding sites
Phyre2
| 16 |
|
PDB 1dgs chain A domain 1
Region: 9 - 60
Aligned: 48
Modelled: 49
Confidence: 6.2%
Identity: 17%
Fold: SAM domain-like
Superfamily: RuvA domain 2-like
Family: NAD+-dependent DNA ligase, domain 3
Phyre2
| 17 |
|
PDB 1rqt chain A
Region: 49 - 57
Aligned: 9
Modelled: 9
Confidence: 6.0%
Identity: 33%
Fold: Ribosomal protein L7/12, oligomerisation (N-terminal) domain
Superfamily: Ribosomal protein L7/12, oligomerisation (N-terminal) domain
Family: Ribosomal protein L7/12, oligomerisation (N-terminal) domain
Phyre2
| 18 |
|
PDB 1rqt chain B
Region: 49 - 57
Aligned: 9
Modelled: 9
Confidence: 6.0%
Identity: 33%
PDB header:ribosome
Chain: B: PDB Molecule:50s ribosomal protein l7/l12;
PDBTitle: nmr structure of dimeric n-terminal domain of ribosomal2 protein l7 from e.coli
Phyre2
| 19 |
|
PDB 1rqt chain A
Region: 49 - 57
Aligned: 9
Modelled: 9
Confidence: 6.0%
Identity: 33%
PDB header:ribosome
Chain: A: PDB Molecule:50s ribosomal protein l7/l12;
PDBTitle: nmr structure of dimeric n-terminal domain of ribosomal2 protein l7 from e.coli
Phyre2
| 20 |
|
PDB 1oyj chain A domain 2
Region: 7 - 21
Aligned: 15
Modelled: 15
Confidence: 5.5%
Identity: 40%
Fold: Thioredoxin fold
Superfamily: Thioredoxin-like
Family: Glutathione S-transferase (GST), N-terminal domain
Phyre2
| 21 |
|
| 22 |
|
| 23 |
|