| 1 |
|
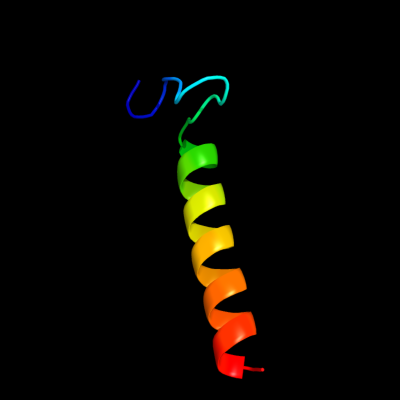
PDB 2i5n chain L domain 1
Region: 12 - 45
Aligned: 34
Modelled: 34
Confidence: 14.0%
Identity: 21%
Fold: Bacterial photosystem II reaction centre, L and M subunits
Superfamily: Bacterial photosystem II reaction centre, L and M subunits
Family: Bacterial photosystem II reaction centre, L and M subunits
Phyre2
| 2 |
|
PDB 2k9y chain B
Region: 266 - 284
Aligned: 19
Modelled: 19
Confidence: 11.3%
Identity: 21%
PDB header:transferase
Chain: B: PDB Molecule:ephrin type-a receptor 2;
PDBTitle: epha2 dimeric structure in the lipidic bicelle at ph 5.0
Phyre2
| 3 |
|
PDB 2j8c chain L domain 1
Region: 11 - 45
Aligned: 35
Modelled: 35
Confidence: 9.6%
Identity: 17%
Fold: Bacterial photosystem II reaction centre, L and M subunits
Superfamily: Bacterial photosystem II reaction centre, L and M subunits
Family: Bacterial photosystem II reaction centre, L and M subunits
Phyre2
| 4 |
|
PDB 2k9y chain A
Region: 266 - 284
Aligned: 19
Modelled: 19
Confidence: 9.1%
Identity: 21%
PDB header:transferase
Chain: A: PDB Molecule:ephrin type-a receptor 2;
PDBTitle: epha2 dimeric structure in the lipidic bicelle at ph 5.0
Phyre2
| 5 |
|
PDB 1eys chain H domain 2
Region: 260 - 288
Aligned: 29
Modelled: 29
Confidence: 7.8%
Identity: 14%
Fold: Single transmembrane helix
Superfamily: Photosystem II reaction centre subunit H, transmembrane region
Family: Photosystem II reaction centre subunit H, transmembrane region
Phyre2
| 6 |
|
PDB 1s7b chain A
Region: 203 - 287
Aligned: 75
Modelled: 84
Confidence: 6.8%
Identity: 15%
Fold: Multidrug resistance efflux transporter EmrE
Superfamily: Multidrug resistance efflux transporter EmrE
Family: Multidrug resistance efflux transporter EmrE
Phyre2
| 7 |
|
PDB 3lrc chain C
Region: 204 - 289
Aligned: 74
Modelled: 86
Confidence: 6.5%
Identity: 14%
PDB header:transport protein
Chain: C: PDB Molecule:arginine/agmatine antiporter;
PDBTitle: structure of e. coli adic (p1)
Phyre2
| 8 |
|
PDB 3rko chain K
Region: 197 - 291
Aligned: 91
Modelled: 92
Confidence: 5.8%
Identity: 15%
PDB header:oxidoreductase
Chain: K: PDB Molecule:nadh-quinone oxidoreductase subunit k;
PDBTitle: crystal structure of the membrane domain of respiratory complex i from2 e. coli at 3.0 angstrom resolution
Phyre2
| 9 |
|
PDB 1k6n chain H
Region: 23 - 55
Aligned: 31
Modelled: 33
Confidence: 5.5%
Identity: 13%
PDB header:photosynthesis
Chain: H: PDB Molecule:photosynthetic reaction center h subunit;
PDBTitle: e(l212)a,d(l213)a double mutant structure of photosynthetic reaction2 center from rhodobacter sphaeroides
Phyre2