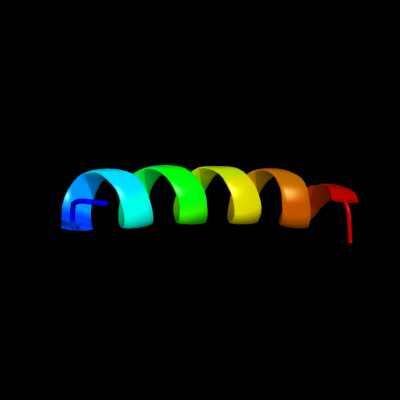| 1 |
|
PDB 3sok chain B
Region: 10 - 30
Aligned: 21
Modelled: 21
Confidence: 28.4%
Identity: 29%
PDB header:cell adhesion
Chain: B: PDB Molecule:fimbrial protein;
PDBTitle: dichelobacter nodosus pilin fima
Phyre2
| 2 |
|
PDB 2ww9 chain B
Region: 21 - 48
Aligned: 25
Modelled: 28
Confidence: 26.6%
Identity: 24%
PDB header:ribosome
Chain: B: PDB Molecule:protein transport protein sss1;
PDBTitle: cryo-em structure of the active yeast ssh1 complex bound to the2 yeast 80s ribosome
Phyre2
| 3 |
|
PDB 1oqw chain A
Region: 10 - 30
Aligned: 21
Modelled: 21
Confidence: 26.4%
Identity: 29%
Fold: Pili subunits
Superfamily: Pili subunits
Family: Pilin
Phyre2
| 4 |
|
PDB 2hg5 chain D
Region: 10 - 34
Aligned: 25
Modelled: 25
Confidence: 15.5%
Identity: 24%
PDB header:membrane protein
Chain: D: PDB Molecule:kcsa channel;
PDBTitle: cs+ complex of a k channel with an amide to ester substitution in the2 selectivity filter
Phyre2
| 5 |
|
PDB 2l0o chain A
Region: 55 - 68
Aligned: 14
Modelled: 14
Confidence: 12.8%
Identity: 21%
PDB header:membrane protein
Chain: A: PDB Molecule:oxidoreductase that catalyzes reoxidation of dsba protein
PDBTitle: dsbb3 peptide structure in 100% tfe
Phyre2
| 6 |
|
PDB 2axt chain J domain 1
Region: 25 - 41
Aligned: 17
Modelled: 17
Confidence: 8.6%
Identity: 24%
Fold: Single transmembrane helix
Superfamily: Photosystem II reaction center protein J, PsbJ
Family: PsbJ-like
Phyre2
| 7 |
|
PDB 2k21 chain A
Region: 4 - 27
Aligned: 18
Modelled: 24
Confidence: 8.0%
Identity: 50%
PDB header:membrane protein
Chain: A: PDB Molecule:potassium voltage-gated channel subfamily e
PDBTitle: nmr structure of human kcne1 in lmpg micelles at ph 6.0 and2 40 degree c
Phyre2
| 8 |
|
PDB 1rhz chain B
Region: 21 - 48
Aligned: 25
Modelled: 25
Confidence: 6.3%
Identity: 16%
Fold: Single transmembrane helix
Superfamily: Preprotein translocase SecE subunit
Family: Preprotein translocase SecE subunit
Phyre2
| 9 |
|
PDB 3mp7 chain B
Region: 35 - 53
Aligned: 19
Modelled: 19
Confidence: 5.6%
Identity: 26%
PDB header:protein transport
Chain: B: PDB Molecule:preprotein translocase subunit sece;
PDBTitle: lateral opening of a translocon upon entry of protein suggests the2 mechanism of insertion into membranes
Phyre2
| 10 |
|
PDB 2jag chain A
Region: 15 - 42
Aligned: 27
Modelled: 28
Confidence: 5.5%
Identity: 26%
PDB header:membrane protein
Chain: A: PDB Molecule:halorhodopsin;
PDBTitle: l1-intermediate of halorhodopsin t203v
Phyre2
| 11 |
|
PDB 3fgx chain A
Region: 64 - 78
Aligned: 15
Modelled: 15
Confidence: 5.3%
Identity: 27%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:rbstp2171;
PDBTitle: structure of uncharacterised protein rbstp2171 from bacillus2 stearothermophilus
Phyre2
| 12 |
|
PDB 3a7k chain D
Region: 15 - 42
Aligned: 27
Modelled: 28
Confidence: 5.3%
Identity: 15%
PDB header:membrane protein
Chain: D: PDB Molecule:halorhodopsin;
PDBTitle: crystal structure of halorhodopsin from natronomonas2 pharaonis
Phyre2