| 1 |
|
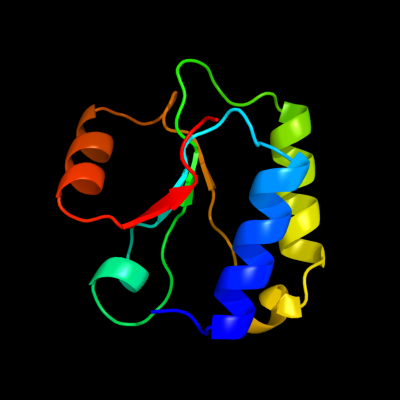
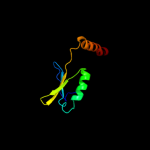
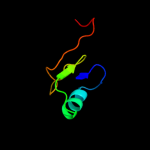
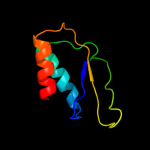
PDB 1no5 chain A
Region: 149 - 244
Aligned: 94
Modelled: 96
Confidence: 40.2%
Identity: 13%
Fold: Nucleotidyltransferase
Superfamily: Nucleotidyltransferase
Family: Catalytic subunit of bi-partite nucleotidyltransferase
Phyre2
| 2 |
|
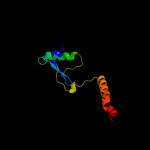
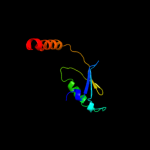
PDB 1r7g chain A
Region: 246 - 252
Aligned: 7
Modelled: 7
Confidence: 29.2%
Identity: 57%
PDB header:membrane protein
Chain: A: PDB Molecule:genome polyprotein;
PDBTitle: nmr structure of the membrane anchor domain (1-31) of the2 nonstructural protein 5a (ns5a) of hepatitis c virus3 (minimized average structure, sample in 100mm dpc)
Phyre2
| 3 |
|
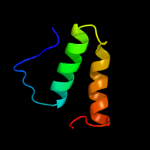
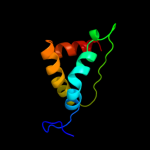
PDB 2l8t chain A
Region: 242 - 258
Aligned: 17
Modelled: 17
Confidence: 20.9%
Identity: 47%
PDB header:structural protein
Chain: A: PDB Molecule:transposon tn557 toxic shock syndrome toxin-1;
PDBTitle: staphylococcus aureus pathogenicity island 1 protein gp6, an internal2 scaffold in size determination
Phyre2
| 4 |
|
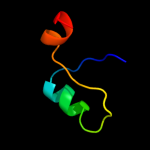
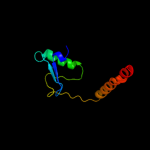
PDB 2aby chain A
Region: 2 - 18
Aligned: 17
Modelled: 17
Confidence: 12.4%
Identity: 29%
PDB header:unknown function
Chain: A: PDB Molecule:hypothetical protein ta0743;
PDBTitle: solution structure of ta0743 from thermoplasma acidophilum
Phyre2
| 5 |
|
PDB 2rff chain A
Region: 154 - 249
Aligned: 90
Modelled: 96
Confidence: 12.1%
Identity: 13%
PDB header:transferase
Chain: A: PDB Molecule:putative nucleotidyltransferase;
PDBTitle: crystal structure of a putative nucleotidyltransferase2 (np_343093.1) from sulfolobus solfataricus at 1.40 a3 resolution
Phyre2
| 6 |
|
PDB 1wot chain A
Region: 152 - 240
Aligned: 85
Modelled: 89
Confidence: 11.8%
Identity: 14%
Fold: Nucleotidyltransferase
Superfamily: Nucleotidyltransferase
Family: Catalytic subunit of bi-partite nucleotidyltransferase
Phyre2
| 7 |
|
PDB 2fok chain A domain 2
Region: 241 - 252
Aligned: 12
Modelled: 12
Confidence: 9.0%
Identity: 42%
Fold: DNA/RNA-binding 3-helical bundle
Superfamily: "Winged helix" DNA-binding domain
Family: Restriction endonuclease FokI, N-terminal (recognition) domain
Phyre2
| 8 |
|
PDB 1dd5 chain A
Region: 88 - 172
Aligned: 78
Modelled: 85
Confidence: 8.6%
Identity: 14%
Fold: RRF/tRNA synthetase additional domain-like
Superfamily: Ribosome recycling factor, RRF
Family: Ribosome recycling factor, RRF
Phyre2
| 9 |
|
PDB 3r1f chain O
Region: 106 - 174
Aligned: 61
Modelled: 69
Confidence: 8.0%
Identity: 18%
PDB header:transcription
Chain: O: PDB Molecule:esx-1 secretion-associated regulator espr;
PDBTitle: crystal structure of a key regulator of virulence in mycobacterium2 tuberculosis
Phyre2
| 10 |
|
PDB 1ge9 chain A
Region: 88 - 172
Aligned: 77
Modelled: 85
Confidence: 7.4%
Identity: 12%
Fold: RRF/tRNA synthetase additional domain-like
Superfamily: Ribosome recycling factor, RRF
Family: Ribosome recycling factor, RRF
Phyre2
| 11 |
|
PDB 1ek8 chain A
Region: 88 - 172
Aligned: 78
Modelled: 85
Confidence: 7.3%
Identity: 15%
Fold: RRF/tRNA synthetase additional domain-like
Superfamily: Ribosome recycling factor, RRF
Family: Ribosome recycling factor, RRF
Phyre2
| 12 |
|
PDB 1a3a chain A
Region: 140 - 187
Aligned: 48
Modelled: 48
Confidence: 6.9%
Identity: 17%
Fold: Phoshotransferase/anion transport protein
Superfamily: Phoshotransferase/anion transport protein
Family: IIA domain of mannitol-specific and ntr phosphotransferase EII
Phyre2
| 13 |
|
PDB 1s6l chain A domain 2
Region: 65 - 96
Aligned: 32
Modelled: 32
Confidence: 6.2%
Identity: 19%
Fold: NosL/MerB-like
Superfamily: NosL/MerB-like
Family: MerB-like
Phyre2
| 14 |
|
PDB 3cu2 chain A
Region: 64 - 109
Aligned: 46
Modelled: 46
Confidence: 6.1%
Identity: 15%
PDB header:isomerase
Chain: A: PDB Molecule:ribulose-5-phosphate 3-epimerase;
PDBTitle: crystal structure of ribulose-5-phosphate 3-epimerase (yp_718263.1)2 from haemophilus somnus 129pt at 1.91 a resolution
Phyre2
| 15 |
|
PDB 1eh1 chain A
Region: 88 - 172
Aligned: 78
Modelled: 85
Confidence: 5.9%
Identity: 9%
Fold: RRF/tRNA synthetase additional domain-like
Superfamily: Ribosome recycling factor, RRF
Family: Ribosome recycling factor, RRF
Phyre2
| 16 |
|
PDB 1sm4 chain A domain 2
Region: 105 - 180
Aligned: 71
Modelled: 76
Confidence: 5.8%
Identity: 10%
Fold: Ferredoxin reductase-like, C-terminal NADP-linked domain
Superfamily: Ferredoxin reductase-like, C-terminal NADP-linked domain
Family: Reductases
Phyre2
| 17 |
|
PDB 1is1 chain A
Region: 88 - 172
Aligned: 78
Modelled: 85
Confidence: 5.8%
Identity: 10%
Fold: RRF/tRNA synthetase additional domain-like
Superfamily: Ribosome recycling factor, RRF
Family: Ribosome recycling factor, RRF
Phyre2
| 18 |
|
PDB 2qmw chain A domain 2
Region: 141 - 210
Aligned: 67
Modelled: 69
Confidence: 5.5%
Identity: 12%
Fold: Ferredoxin-like
Superfamily: ACT-like
Family: Phenylalanine metabolism regulatory domain
Phyre2
| 19 |
|
PDB 3gny chain A
Region: 240 - 248
Aligned: 9
Modelled: 9
Confidence: 5.4%
Identity: 33%
PDB header:antimicrobial protein
Chain: A: PDB Molecule:neutrophil defensin 1;
PDBTitle: crystal structure of human alpha-defensin 1 (hnp1)
Phyre2
| 20 |
|
PDB 3hj2 chain B
Region: 240 - 248
Aligned: 9
Modelled: 9
Confidence: 5.2%
Identity: 33%
PDB header:antimicrobial protein
Chain: B: PDB Molecule:human neutrophil peptide 1;
PDBTitle: crystal structure of covalent dimer of hnp1
Phyre2
| 21 |
|