| 1 |
|
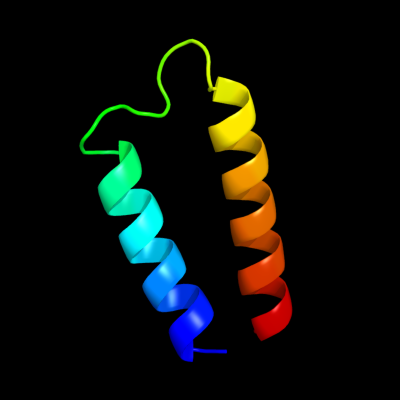
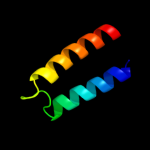
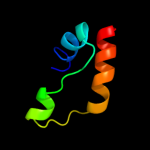
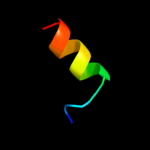
PDB 1q90 chain B
Region: 110 - 153
Aligned: 44
Modelled: 44
Confidence: 36.6%
Identity: 18%
Fold: Heme-binding four-helical bundle
Superfamily: Transmembrane di-heme cytochromes
Family: Cytochrome b of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)
Phyre2
| 2 |
|
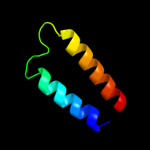
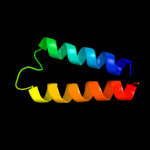
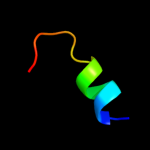
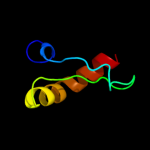
PDB 2e74 chain A domain 1
Region: 110 - 153
Aligned: 44
Modelled: 44
Confidence: 31.9%
Identity: 20%
Fold: Heme-binding four-helical bundle
Superfamily: Transmembrane di-heme cytochromes
Family: Cytochrome b of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)
Phyre2
| 3 |
|
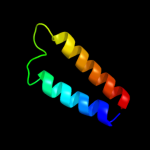
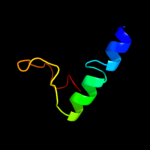
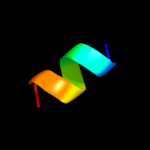
PDB 3cx5 chain C domain 2
Region: 110 - 153
Aligned: 44
Modelled: 44
Confidence: 24.2%
Identity: 11%
Fold: Heme-binding four-helical bundle
Superfamily: Transmembrane di-heme cytochromes
Family: Cytochrome b of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)
Phyre2
| 4 |
|
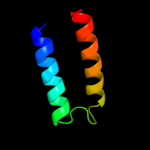
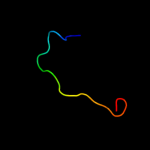
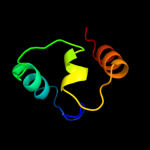
PDB 3cx5 chain N
Region: 110 - 153
Aligned: 44
Modelled: 44
Confidence: 16.8%
Identity: 11%
PDB header:oxidoreductase
Chain: N: PDB Molecule:cytochrome b;
PDBTitle: structure of complex iii with bound cytochrome c in reduced2 state and definition of a minimal core interface for3 electron transfer.
Phyre2
| 5 |
|
PDB 2qjk chain M
Region: 110 - 153
Aligned: 44
Modelled: 44
Confidence: 16.0%
Identity: 14%
PDB header:electron transport
Chain: M: PDB Molecule:cytochrome b;
PDBTitle: crystal structure analysis of mutant rhodobacter2 sphaeroides bc1 with stigmatellin and antimycin
Phyre2
| 6 |
|
PDB 2au5 chain A domain 1
Region: 59 - 98
Aligned: 38
Modelled: 40
Confidence: 12.2%
Identity: 18%
Fold: EF2947-like
Superfamily: EF2947-like
Family: EF2947-like
Phyre2
| 7 |
|
PDB 1p4q chain A
Region: 85 - 103
Aligned: 19
Modelled: 19
Confidence: 11.0%
Identity: 26%
PDB header:transcription/transferase
Chain: A: PDB Molecule:cbp/p300-interacting transactivator 2;
PDBTitle: solution structure of the cited2 transactivation domain in2 complex with the p300 ch1 domain
Phyre2
| 8 |
|
PDB 2vkj chain A
Region: 72 - 115
Aligned: 37
Modelled: 44
Confidence: 10.1%
Identity: 43%
PDB header:membrane protein
Chain: A: PDB Molecule:tm1634;
PDBTitle: structure of the soluble domain of the membrane protein2 tm1634 from thermotoga maritima
Phyre2
| 9 |
|
PDB 2yv4 chain A
Region: 150 - 164
Aligned: 14
Modelled: 15
Confidence: 9.0%
Identity: 14%
PDB header:rna binding protein
Chain: A: PDB Molecule:hypothetical protein ph0435;
PDBTitle: crystal structure of c-terminal sua5 domain from pyrococcus horikoshii2 hypothetical sua5 protein ph0435
Phyre2
| 10 |
|
PDB 2p62 chain A domain 1
Region: 76 - 84
Aligned: 9
Modelled: 9
Confidence: 8.3%
Identity: 22%
Fold: PH0156-like
Superfamily: PH0156-like
Family: PH0156-like
Phyre2
| 11 |
|
PDB 2hkj chain A domain 1
Region: 93 - 143
Aligned: 50
Modelled: 51
Confidence: 8.3%
Identity: 14%
Fold: S13-like H2TH domain
Superfamily: S13-like H2TH domain
Family: Topoisomerase VI-B subunit middle domain
Phyre2
| 12 |
|
PDB 1r8u chain A
Region: 85 - 103
Aligned: 19
Modelled: 19
Confidence: 8.1%
Identity: 26%
PDB header:transcription/transcription activator
Chain: A: PDB Molecule:cbp/p300-interacting transactivator 2;
PDBTitle: nmr structure of cbp taz1/cited2 complex
Phyre2
| 13 |
|
PDB 1ppj chain C domain 2
Region: 110 - 153
Aligned: 42
Modelled: 44
Confidence: 7.6%
Identity: 14%
Fold: Heme-binding four-helical bundle
Superfamily: Transmembrane di-heme cytochromes
Family: Cytochrome b of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)
Phyre2
| 14 |
|
PDB 2e9x chain B domain 1
Region: 94 - 108
Aligned: 15
Modelled: 15
Confidence: 7.3%
Identity: 7%
Fold: GINS helical bundle-like
Superfamily: GINS helical bundle-like
Family: PSF2 C-terminal domain-like
Phyre2
| 15 |
|
PDB 2wp0 chain C
Region: 78 - 124
Aligned: 47
Modelled: 47
Confidence: 7.3%
Identity: 19%
PDB header:dna binding protein
Chain: C: PDB Molecule:chromosomal replication initiator protein dnaa;
PDBTitle: crystal structure of a hoba-dnaa (domain i-ii) complex from2 helicobacter pylori.
Phyre2