| 1 |
|
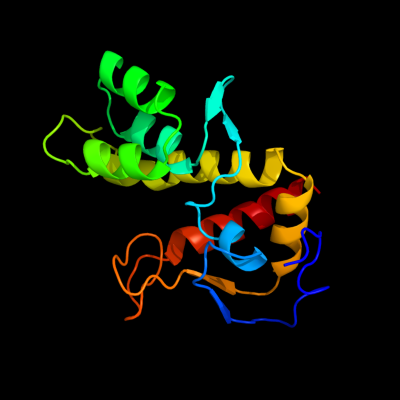
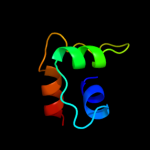
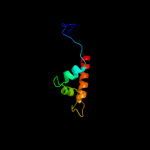
PDB 2b3w chain A domain 1
Region: 1 - 160
Aligned: 160
Modelled: 160
Confidence: 100.0%
Identity: 100%
Fold: YbiA-like
Superfamily: YbiA-like
Family: YbiA-like
Phyre2
| 2 |
|
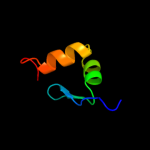
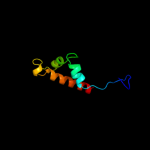
PDB 2k0m chain A
Region: 36 - 84
Aligned: 48
Modelled: 49
Confidence: 45.3%
Identity: 17%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: solution nmr structure of the uncharacterized protein from2 rhodospirillum rubrum gene locus rru_a0810. northeast3 structural genomics target rrr43
Phyre2
| 3 |
|
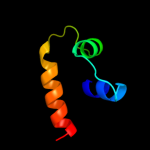
PDB 1w7c chain A domain 3
Region: 30 - 58
Aligned: 29
Modelled: 29
Confidence: 21.7%
Identity: 17%
Fold: Cystatin-like
Superfamily: Amine oxidase N-terminal region
Family: Amine oxidase N-terminal region
Phyre2
| 4 |
|
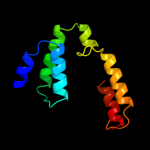
PDB 2ff4 chain A domain 1
Region: 110 - 158
Aligned: 49
Modelled: 49
Confidence: 17.1%
Identity: 14%
Fold: DNA/RNA-binding 3-helical bundle
Superfamily: C-terminal effector domain of the bipartite response regulators
Family: PhoB-like
Phyre2
| 5 |
|
PDB 2oar chain A
Region: 147 - 159
Aligned: 13
Modelled: 13
Confidence: 11.7%
Identity: 46%
PDB header:membrane protein
Chain: A: PDB Molecule:large-conductance mechanosensitive channel;
PDBTitle: mechanosensitive channel of large conductance (mscl)
Phyre2
| 6 |
|
PDB 1uur chain A domain 2
Region: 119 - 153
Aligned: 26
Modelled: 35
Confidence: 10.7%
Identity: 19%
Fold: Common fold of diphtheria toxin/transcription factors/cytochrome f
Superfamily: p53-like transcription factors
Family: STAT DNA-binding domain
Phyre2
| 7 |
|
PDB 1avo chain D
Region: 44 - 93
Aligned: 48
Modelled: 50
Confidence: 10.1%
Identity: 19%
PDB header:proteasome activator
Chain: D: PDB Molecule:11s regulator;
PDBTitle: proteasome activator reg(alpha)
Phyre2
| 8 |
|
PDB 1gxq chain A
Region: 110 - 158
Aligned: 48
Modelled: 49
Confidence: 8.9%
Identity: 13%
Fold: DNA/RNA-binding 3-helical bundle
Superfamily: C-terminal effector domain of the bipartite response regulators
Family: PhoB-like
Phyre2
| 9 |
|
PDB 2k4j chain A
Region: 110 - 158
Aligned: 48
Modelled: 49
Confidence: 8.5%
Identity: 10%
PDB header:transcription
Chain: A: PDB Molecule:putative transcriptional regulator;
PDBTitle: arsr dna binding domain
Phyre2
| 10 |
|
PDB 1opc chain A
Region: 110 - 156
Aligned: 46
Modelled: 47
Confidence: 7.5%
Identity: 11%
Fold: DNA/RNA-binding 3-helical bundle
Superfamily: C-terminal effector domain of the bipartite response regulators
Family: PhoB-like
Phyre2
| 11 |
|
PDB 1i3j chain A
Region: 31 - 71
Aligned: 41
Modelled: 41
Confidence: 7.4%
Identity: 10%
Fold: DNA-binding domain of intron-encoded endonucleases
Superfamily: DNA-binding domain of intron-encoded endonucleases
Family: DNA-binding domain of intron-encoded endonucleases
Phyre2
| 12 |
|
PDB 1kgs chain A domain 1
Region: 110 - 158
Aligned: 43
Modelled: 49
Confidence: 7.2%
Identity: 12%
Fold: DNA/RNA-binding 3-helical bundle
Superfamily: C-terminal effector domain of the bipartite response regulators
Family: PhoB-like
Phyre2
| 13 |
|
PDB 1mjd chain A
Region: 6 - 53
Aligned: 42
Modelled: 48
Confidence: 6.4%
Identity: 12%
Fold: beta-Grasp (ubiquitin-like)
Superfamily: Doublecortin (DC)
Family: Doublecortin (DC)
Phyre2
| 14 |
|
PDB 1mhy chain G
Region: 59 - 153
Aligned: 95
Modelled: 95
Confidence: 6.2%
Identity: 21%
Fold: Open three-helical up-and-down bundle
Superfamily: Methane monooxygenase hydrolase, gamma subunit
Family: Methane monooxygenase hydrolase, gamma subunit
Phyre2
| 15 |
|
PDB 2l2l chain A
Region: 147 - 160
Aligned: 14
Modelled: 14
Confidence: 6.1%
Identity: 29%
PDB header:transferase
Chain: A: PDB Molecule:transcriptional repressor p66-alpha;
PDBTitle: solution structure of the coiled-coil complex between mbd2 and2 p66alpha
Phyre2
| 16 |
|
PDB 1mjt chain A
Region: 37 - 108
Aligned: 71
Modelled: 72
Confidence: 5.8%
Identity: 15%
Fold: Nitric oxide (NO) synthase oxygenase domain
Superfamily: Nitric oxide (NO) synthase oxygenase domain
Family: Nitric oxide (NO) synthase oxygenase domain
Phyre2
| 17 |
|
PDB 1om4 chain A
Region: 37 - 108
Aligned: 71
Modelled: 72
Confidence: 5.3%
Identity: 15%
Fold: Nitric oxide (NO) synthase oxygenase domain
Superfamily: Nitric oxide (NO) synthase oxygenase domain
Family: Nitric oxide (NO) synthase oxygenase domain
Phyre2