| 1 |
|
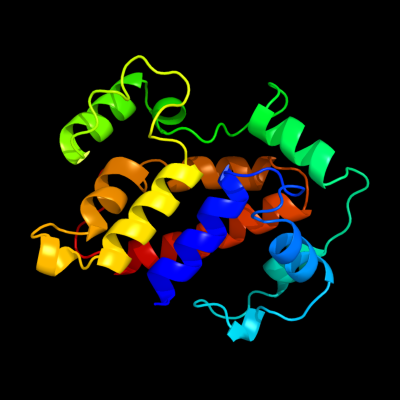
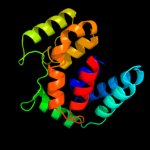
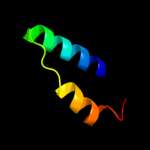
PDB 1s9u chain A
Region: 2 - 184
Aligned: 177
Modelled: 183
Confidence: 100.0%
Identity: 32%
Fold: TorD-like
Superfamily: TorD-like
Family: TorD-like
Phyre2
| 2 |
|
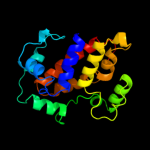
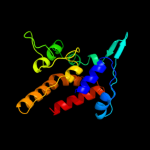
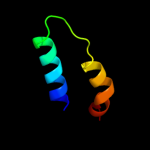
PDB 1n1c chain A
Region: 1 - 183
Aligned: 179
Modelled: 183
Confidence: 100.0%
Identity: 19%
Fold: TorD-like
Superfamily: TorD-like
Family: TorD-like
Phyre2
| 3 |
|
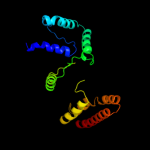
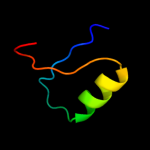
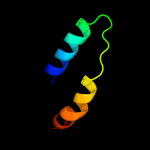
PDB 2o9x chain A
Region: 5 - 172
Aligned: 143
Modelled: 154
Confidence: 100.0%
Identity: 24%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:reductase, assembly protein;
PDBTitle: crystal structure of a putative redox enzyme maturation protein from2 archaeoglobus fulgidus
Phyre2
| 4 |
|
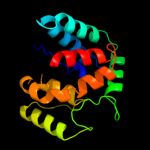
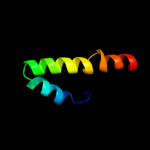
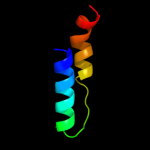
PDB 2o9x chain A domain 1
Region: 7 - 172
Aligned: 141
Modelled: 153
Confidence: 100.0%
Identity: 23%
Fold: TorD-like
Superfamily: TorD-like
Family: TorD-like
Phyre2
| 5 |
|
PDB 2idg chain A domain 1
Region: 4 - 167
Aligned: 140
Modelled: 158
Confidence: 99.8%
Identity: 19%
Fold: TorD-like
Superfamily: TorD-like
Family: TorD-like
Phyre2
| 6 |
|
PDB 3bf2 chain A
Region: 77 - 108
Aligned: 31
Modelled: 32
Confidence: 22.3%
Identity: 26%
PDB header:lipoprotein
Chain: A: PDB Molecule:putative lipoprotein;
PDBTitle: crystal structure of the a1ksw9_neimf protein from2 neisseria meningitidis. northeast structural genomics3 consortium target mr36a
Phyre2
| 7 |
|
PDB 2zop chain A
Region: 108 - 153
Aligned: 44
Modelled: 46
Confidence: 14.4%
Identity: 11%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:putative uncharacterized protein tthb164;
PDBTitle: x-ray crystal structure of a crispr-associated cmr5 family2 protein from thermus thermophilus hb8
Phyre2
| 8 |
|
PDB 1ysg chain A domain 1
Region: 109 - 146
Aligned: 38
Modelled: 38
Confidence: 13.3%
Identity: 21%
Fold: Toxins' membrane translocation domains
Superfamily: Bcl-2 inhibitors of programmed cell death
Family: Bcl-2 inhibitors of programmed cell death
Phyre2
| 9 |
|
PDB 1q8h chain A
Region: 105 - 113
Aligned: 9
Modelled: 9
Confidence: 9.6%
Identity: 56%
Fold: GLA-domain
Superfamily: GLA-domain
Family: GLA-domain
Phyre2
| 10 |
|
PDB 1q8h chain A
Region: 105 - 113
Aligned: 9
Modelled: 9
Confidence: 9.6%
Identity: 56%
PDB header:metal binding protein
Chain: A: PDB Molecule:osteocalcin;
PDBTitle: crystal structure of porcine osteocalcin
Phyre2
| 11 |
|
PDB 3qbr chain A
Region: 109 - 144
Aligned: 36
Modelled: 36
Confidence: 9.1%
Identity: 14%
PDB header:apoptosis
Chain: A: PDB Molecule:sjchgc06286 protein;
PDBTitle: bakbh3 in complex with sja
Phyre2
| 12 |
|
PDB 1f16 chain A
Region: 109 - 146
Aligned: 38
Modelled: 38
Confidence: 8.5%
Identity: 24%
Fold: Toxins' membrane translocation domains
Superfamily: Bcl-2 inhibitors of programmed cell death
Family: Bcl-2 inhibitors of programmed cell death
Phyre2
| 13 |
|
PDB 2pon chain B domain 1
Region: 109 - 146
Aligned: 38
Modelled: 38
Confidence: 8.5%
Identity: 21%
Fold: Toxins' membrane translocation domains
Superfamily: Bcl-2 inhibitors of programmed cell death
Family: Bcl-2 inhibitors of programmed cell death
Phyre2
| 14 |
|
PDB 1g5m chain A
Region: 110 - 146
Aligned: 37
Modelled: 37
Confidence: 7.5%
Identity: 14%
Fold: Toxins' membrane translocation domains
Superfamily: Bcl-2 inhibitors of programmed cell death
Family: Bcl-2 inhibitors of programmed cell death
Phyre2
| 15 |
|
PDB 1pq1 chain A
Region: 109 - 146
Aligned: 38
Modelled: 38
Confidence: 7.3%
Identity: 21%
Fold: Toxins' membrane translocation domains
Superfamily: Bcl-2 inhibitors of programmed cell death
Family: Bcl-2 inhibitors of programmed cell death
Phyre2
| 16 |
|
PDB 2vof chain A
Region: 109 - 146
Aligned: 38
Modelled: 38
Confidence: 7.1%
Identity: 13%
PDB header:apoptosis
Chain: A: PDB Molecule:bcl-2-related protein a1;
PDBTitle: structure of mouse a1 bound to the puma bh3-domain
Phyre2
| 17 |
|
PDB 1q3m chain A
Region: 105 - 113
Aligned: 9
Modelled: 9
Confidence: 6.9%
Identity: 56%
Fold: GLA-domain
Superfamily: GLA-domain
Family: GLA-domain
Phyre2
| 18 |
|
PDB 1rut chain X domain 2
Region: 61 - 68
Aligned: 8
Modelled: 8
Confidence: 6.4%
Identity: 50%
Fold: Glucocorticoid receptor-like (DNA-binding domain)
Superfamily: Glucocorticoid receptor-like (DNA-binding domain)
Family: LIM domain
Phyre2
| 19 |
|
PDB 1m3v chain A domain 2
Region: 61 - 68
Aligned: 8
Modelled: 8
Confidence: 6.2%
Identity: 50%
Fold: Glucocorticoid receptor-like (DNA-binding domain)
Superfamily: Glucocorticoid receptor-like (DNA-binding domain)
Family: LIM domain
Phyre2
| 20 |
|
PDB 1zy3 chain A domain 1
Region: 109 - 146
Aligned: 38
Modelled: 38
Confidence: 6.0%
Identity: 18%
Fold: Toxins' membrane translocation domains
Superfamily: Bcl-2 inhibitors of programmed cell death
Family: Bcl-2 inhibitors of programmed cell death
Phyre2
| 21 |
|
| 22 |
|
| 23 |
|