| Secondary structure and disorder prediction | |
 |
| | |
1 | . | . | . | . | . | . | . | . | 10 | . | . | . | . | . | . | . | . | . | 20 | . | . | . | . | . | . | . | . | . | 30 | . | . | . | . | . | . | . | . | . | 40 | . | . | . | . | . | . | . | . | . | 50 | . | . | . | . | . | . | . | . | . | 60 |
| Sequence | |
M | S | N | A | Q | E | A | V | K | T | R | H | K | E | T | S | L | I | F | P | V | L | A | L | V | V | L | F | L | W | G | S | S | Q | T | L | P | V | V | I | A | I | N | L | L | A | L | I | G | I | L | S | S | A | F | S | V | V | R | H |
| Secondary structure | |
|
|
|
|  |  |  |  |
|
|  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |
|
|
|
|
|
|  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |
| SS confidence | |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| Disorder | |
? | ? | ? | ? | ? | ? | ? | ? | ? | ? |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| ? |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| Disorder confidence | |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| |
| | |
. | . | . | . | . | . | . | . | . | 70 | . | . | . | . | . | . | . | . | . | 80 | . | . | . | . | . | . | . | . | . | 90 | . | . | . | . | . | . | . | . | . | 100 | . | . | . | . | . | . | . | . | . | 110 | . | . | . | . | . | . | . | . | . | 120 |
| Sequence | |
A | D | V | L | A | H | R | L | G | E | P | Y | G | S | L | I | L | S | L | S | V | V | I | L | E | V | S | L | I | S | A | L | M | A | T | G | D | A | A | P | T | L | M | R | D | T | L | Y | S | I | I | M | I | V | T | G | G | L | V | G |
| Secondary structure | |
 |  |  |  |  |  |  |  |
|
|
|  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |
|
|
|
|
|  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |
| SS confidence | |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| Disorder | |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| ? | ? | ? | ? | ? |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| Disorder confidence | |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| |
| | |
. | . | . | . | . | . | . | . | . | 130 | . | . | . | . | . | . | . | . | . | 140 | . | . | . | . | . | . | . | . | . | 150 | . | . | . | . | . | . | . | . | . | 160 | . | . | . | . | . | . | . | . | . | 170 | . | . | . | . | . | . | . | . | . | 180 |
| Sequence | |
F | S | L | L | L | G | G | R | K | F | A | T | Q | Y | M | N | L | F | G | I | K | Q | Y | L | I | A | L | F | P | L | A | I | I | V | L | V | F | P | M | A | L | P | A | A | N | F | S | T | G | Q | A | L | L | V | A | L | I | S | A | A |
| Secondary structure | |
 |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |
|
|
|
|
|
|
|  |  |  |  |  |  |  |  |  |  |  |  |  |
| SS confidence | |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| Disorder | |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| ? |
|
| ? | ? | ? | ? | ? | ? | ? |
| ? |
|
|
|
|
|
|
|
|
|
|
|
|
| Disorder confidence | |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| |
| | |
. | . | . | . | . | . | . | . | . | 190 | . | . | . | . | . | . | . | . | . | 200 | . | . | . | . | . | . | . | . | . | 210 | . | . | . | . | . | . | . | . | . | 220 | . | . | . | . | . | . | . | . | . | 230 | . | . | . | . | . | . | . | . | . | 240 |
| Sequence | |
M | Y | G | V | F | L | L | I | Q | T | K | T | H | Q | S | L | F | V | Y | E | H | E | D | D | S | D | D | D | D | P | H | H | G | K | P | S | A | H | S | S | L | W | H | A | I | W | L | I | I | H | L | I | A | V | I | A | V | T | K | M |
| Secondary structure | |
 |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |
| SS confidence | |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| Disorder | |
|
|
|
|
|
|
|
|
|
|
| ? | ? | ? | ? | ? | ? | ? | ? | ? | ? | ? | ? | ? | ? | ? | ? | ? | ? | ? | ? | ? | ? | ? | ? | ? | ? |
|
| ? |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| Disorder confidence | |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| |
| | |
. | . | . | . | . | . | . | . | . | 250 | . | . | . | . | . | . | . | . | . | 260 | . | . | . | . | . | . | . | . | . | 270 | . | . | . | . | . | . | . | . | . | 280 | . | . | . | . | . | . | . | . | . | 290 | . | . | . | . | . | . | . | . | . | 300 |
| Sequence | |
N | A | S | S | L | E | T | L | L | D | S | M | N | A | P | V | A | F | T | G | F | L | V | A | L | L | I | L | S | P | E | G | L | G | A | L | K | A | V | L | N | N | Q | V | Q | R | A | M | N | L | F | F | G | S | V | L | A | T | I | S |
| Secondary structure | |
 |  |  |  |  |  |  |  |  |  |  |
|
|
|
|  |  |  |  |  |  |  |  |  |  |  |
|
|
|  |  |  |  |  |  |  |  |  |  |  |
|
|
|  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |
| SS confidence | |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| Disorder | |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| Disorder confidence | |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| |
| | |
. | . | . | . | . | . | . | . | . | 310 | . | . | . | . | . | . | . | . | . | 320 | . | . | . | . | . | . | . | . | . | 330 | . | . | . | . | . | . | . | . | . | 340 | . | . | . | . | . | . | . | . | . | 350 | . | . | . | . | . | . | . | . | . | 360 |
| Sequence | |
L | T | V | P | V | V | T | L | I | A | F | M | T | G | N | E | L | Q | F | A | L | G | A | P | E | M | V | V | M | V | A | S | L | V | L | C | H | I | S | F | S | T | G | R | T | N | V | L | N | G | A | A | H | L | A | L | F | A | A | Y |
| Secondary structure | |
 |  |  |  |  |  |  |  |  |  |  |  |  |
|
|
|
|
|
|
|
|
|  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |
|
|
|
|
|
|  |  |  |  |  |  |  |  |  |  |  |  |  |  |
| SS confidence | |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| Disorder | |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| Disorder confidence | |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| |
| | |
. | . | . | . | . | . |
| Sequence | |
L | M | T | I | F | A |
| Secondary structure | |
 |  |  |  |
|
|
| SS confidence | |
|
|
|
|
|
|
| Disorder | |
|
|
|
| ? | ? |
| Disorder confidence | |
|
|
|
|
|
|
| |
| Confidence Key |
| High(9) | |
|
|
|
|
|
|
|
|
|
Low (0) |
| ? | Disordered |
  | Alpha helix |
  | Beta strand |
Hover over an aligned region to see model and summary info
Please note, only up to the top 20 hits are modelled to reduce computer load
|
| 1 |
|
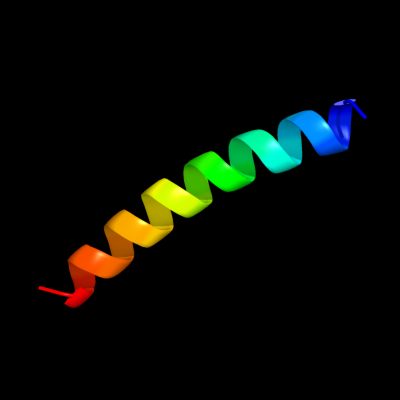
PDB 1xrd chain A domain 1
Region: 323 - 350
Aligned: 28
Modelled: 28
Confidence: 55.8%
Identity: 32%
Fold: Light-harvesting complex subunits
Superfamily: Light-harvesting complex subunits
Family: Light-harvesting complex subunits
Phyre2
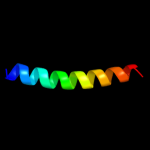
| 2 |
|
PDB 3rko chain M
Region: 111 - 252
Aligned: 139
Modelled: 142
Confidence: 24.2%
Identity: 9%
PDB header:oxidoreductase
Chain: M: PDB Molecule:nadh-quinone oxidoreductase subunit m;
PDBTitle: crystal structure of the membrane domain of respiratory complex i from2 e. coli at 3.0 angstrom resolution
Phyre2
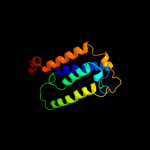
| 3 |
|
PDB 2oar chain A domain 1
Region: 279 - 310
Aligned: 32
Modelled: 32
Confidence: 23.3%
Identity: 22%
Fold: Gated mechanosensitive channel
Superfamily: Gated mechanosensitive channel
Family: Gated mechanosensitive channel
Phyre2
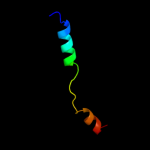
| 4 |
|
PDB 2oar chain A
Region: 277 - 310
Aligned: 34
Modelled: 34
Confidence: 15.9%
Identity: 18%
PDB header:membrane protein
Chain: A: PDB Molecule:large-conductance mechanosensitive channel;
PDBTitle: mechanosensitive channel of large conductance (mscl)
Phyre2
| 5 |
|
PDB 3mk7 chain F
Region: 171 - 236
Aligned: 66
Modelled: 66
Confidence: 12.2%
Identity: 6%
PDB header:oxidoreductase
Chain: F: PDB Molecule:cytochrome c oxidase, cbb3-type, subunit p;
PDBTitle: the structure of cbb3 cytochrome oxidase
Phyre2
| 6 |
|
PDB 1twf chain C domain 2
Region: 62 - 69
Aligned: 8
Modelled: 8
Confidence: 7.8%
Identity: 63%
Fold: Insert subdomain of RNA polymerase alpha subunit
Superfamily: Insert subdomain of RNA polymerase alpha subunit
Family: Insert subdomain of RNA polymerase alpha subunit
Phyre2
| 7 |
|
PDB 1wgl chain A
Region: 270 - 293
Aligned: 24
Modelled: 24
Confidence: 6.3%
Identity: 13%
Fold: RuvA C-terminal domain-like
Superfamily: UBA-like
Family: CUE domain
Phyre2
|
| Detailed template information | |
Due to computational demand, binding site predictions are not run for batch jobs
If you want to predict binding sites, please manually submit your model of choice to 3DLigandSite
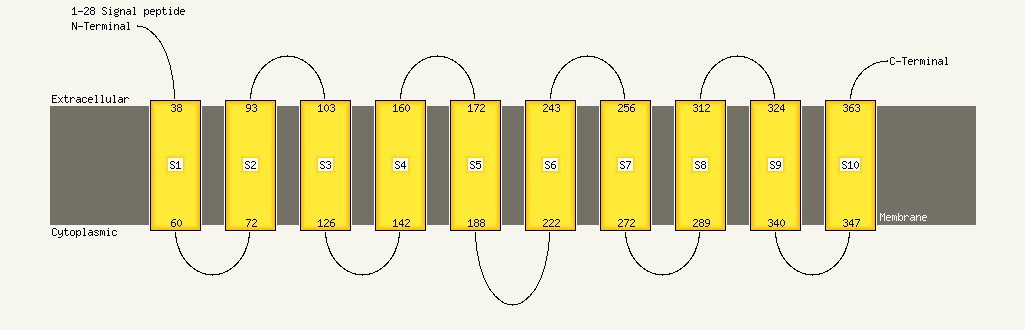
| Transmembrane helix prediction | |
Transmembrane helices have been predicted in your sequence to adopt the topology shown below
Phyre is for academic use only
| Please cite: Protein structure prediction on
the web: a case study using the Phyre server |
| Kelley LA and Sternberg MJE. Nature Protocols
4, 363 - 371 (2009) [pdf] [Import into BibTeX] |
| |
| If you use the binding site
predictions from 3DLigandSite, please also cite: |
| 3DLigandSite: predicting ligand-binding sites using similar structures. |
| Wass MN, Kelley LA and Sternberg
MJ Nucleic Acids Research 38, W469-73 (2010) [PubMed] |
| |
|
|
|
|