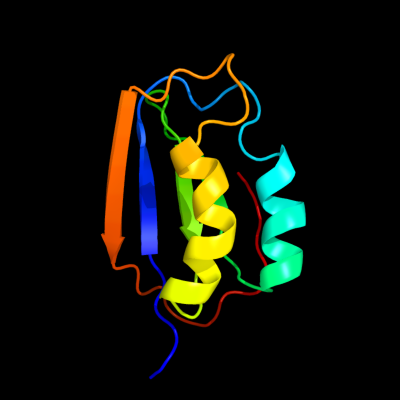
| 1 | c2gv1A_
|
|
 |
100.0 |
100 |
PDB header:hydrolase
Chain: A: PDB Molecule:probable acylphosphatase;
PDBTitle: nmr solution structure of the acylphosphatase from2 eschaerichia coli
|
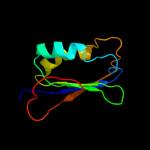
| 2 | c3br8A_
|
|
 |
100.0 |
36 |
PDB header:hydrolase
Chain: A: PDB Molecule:probable acylphosphatase;
PDBTitle: crystal structure of acylphosphatase from bacillus subtilis
|
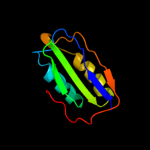
| 3 | c2bjeA_
|
|
 |
100.0 |
51 |
PDB header:hydrolase
Chain: A: PDB Molecule:acylphosphatase;
PDBTitle: acylphosphatase from sulfolobus solfataricus. monclinic p212 space group
|
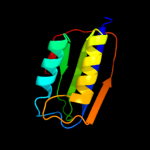
| 4 | d2acya_
|
|
 |
100.0 |
32 |
Fold:Ferredoxin-like
Superfamily:Acylphosphatase/BLUF domain-like
Family:Acylphosphatase-like |
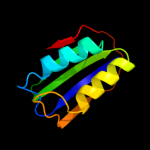
| 5 | d1urra_
|
|
 |
100.0 |
30 |
Fold:Ferredoxin-like
Superfamily:Acylphosphatase/BLUF domain-like
Family:Acylphosphatase-like |
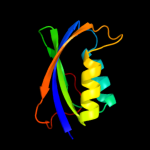
| 6 | d1w2ia_
|
|
 |
100.0 |
48 |
Fold:Ferredoxin-like
Superfamily:Acylphosphatase/BLUF domain-like
Family:Acylphosphatase-like |
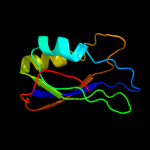
| 7 | d1apsa_
|
|
 |
100.0 |
33 |
Fold:Ferredoxin-like
Superfamily:Acylphosphatase/BLUF domain-like
Family:Acylphosphatase-like |
| 8 | d1ulra_
|
|
 |
100.0 |
49 |
Fold:Ferredoxin-like
Superfamily:Acylphosphatase/BLUF domain-like
Family:Acylphosphatase-like |
| 9 | d1gxua_
|
|
 |
100.0 |
36 |
Fold:Ferredoxin-like
Superfamily:Acylphosphatase/BLUF domain-like
Family:Acylphosphatase-like |
| 10 | d2buna1
|
|
 |
91.2 |
19 |
Fold:Ferredoxin-like
Superfamily:Acylphosphatase/BLUF domain-like
Family:BLUF domain |
| 11 | d1yrxa1
|
|
 |
90.6 |
21 |
Fold:Ferredoxin-like
Superfamily:Acylphosphatase/BLUF domain-like
Family:BLUF domain |
| 12 | c2hfnJ_
|
|
 |
89.6 |
16 |
PDB header:electron transport
Chain: J: PDB Molecule:synechocystis photoreceptor (slr1694);
PDBTitle: crystal structures of the synechocystis photoreceptor slr1694 reveal2 distinct structural states related to signaling
|
| 13 | c2kb2A_
|
|
 |
89.5 |
21 |
PDB header:signaling protein, hydrolase regulator
Chain: A: PDB Molecule:blrp1;
PDBTitle: blrp1 bluf
|
| 14 | d2byca1
|
|
 |
88.1 |
18 |
Fold:Ferredoxin-like
Superfamily:Acylphosphatase/BLUF domain-like
Family:BLUF domain |
| 15 | d1x0pa1
|
|
 |
87.5 |
18 |
Fold:Ferredoxin-like
Superfamily:Acylphosphatase/BLUF domain-like
Family:BLUF domain |
| 16 | c2xznF_
|
|
 |
85.8 |
14 |
PDB header:ribosome
Chain: F: PDB Molecule:eif1;
PDBTitle: crystal structure of the eukaryotic 40s ribosomal2 subunit in complex with initiation factor 1. this file3 contains the 40s subunit and initiation factor for4 molecule 2
|
| 17 | d2qswa1
|
|
 |
85.4 |
15 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:NIL domain-like |
| 18 | c2oghA_
|
|
 |
81.2 |
11 |
PDB header:translation
Chain: A: PDB Molecule:eukaryotic translation initiation factor eif-1;
PDBTitle: solution structure of yeast eif1
|
| 19 | d2if1a_
|
|
 |
79.7 |
9 |
Fold:eIF1-like
Superfamily:eIF1-like
Family:eIF1-like |
| 20 | d2f1fa2
|
|
 |
78.2 |
12 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:IlvH-like |
| 21 | d2fgca1 |
|
not modelled |
76.4 |
12 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:IlvH-like |
| 22 | d2pc6a1 |
|
not modelled |
74.9 |
12 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:IlvH-like |
| 23 | c2fgcA_ |
|
not modelled |
64.8 |
12 |
PDB header:transferase
Chain: A: PDB Molecule:acetolactate synthase, small subunit;
PDBTitle: crystal structure of acetolactate synthase- small subunit from2 thermotoga maritima
|
| 24 | c3c5tB_ |
|
not modelled |
64.5 |
53 |
PDB header:signaling protein/signaling protein
Chain: B: PDB Molecule:exendin-4;
PDBTitle: crystal structure of the ligand-bound glucagon-like peptide-1 receptor2 extracellular domain
|
| 25 | c2pc6C_ |
|
not modelled |
61.8 |
11 |
PDB header:lyase
Chain: C: PDB Molecule:probable acetolactate synthase isozyme iii (small subunit);
PDBTitle: crystal structure of putative acetolactate synthase- small subunit2 from nitrosomonas europaea
|
| 26 | c2f1fA_ |
|
not modelled |
61.0 |
12 |
PDB header:transferase
Chain: A: PDB Molecule:acetolactate synthase isozyme iii small subunit;
PDBTitle: crystal structure of the regulatory subunit of2 acetohydroxyacid synthase isozyme iii from e. coli
|
| 27 | c2rjzA_ |
|
not modelled |
58.3 |
14 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:pilo protein;
PDBTitle: crystal structure of the type 4 fimbrial biogenesis protein pilo from2 pseudomonas aeruginosa
|
| 28 | d3dhxa1 |
|
not modelled |
56.6 |
15 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:NIL domain-like |
| 29 | d2qrra1 |
|
not modelled |
56.1 |
13 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:NIL domain-like |
| 30 | d1a0ia1 |
|
not modelled |
40.6 |
16 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:DNA ligase/mRNA capping enzyme postcatalytic domain |
| 31 | d2zjre1 |
|
not modelled |
25.1 |
16 |
Fold:Ribosomal protein L6
Superfamily:Ribosomal protein L6
Family:Ribosomal protein L6 |
| 32 | c3fhaD_ |
|
not modelled |
23.6 |
18 |
PDB header:hydrolase
Chain: D: PDB Molecule:endo-beta-n-acetylglucosaminidase;
PDBTitle: structure of endo-beta-n-acetylglucosaminidase a
|
| 33 | c2yy3B_ |
|
not modelled |
22.7 |
21 |
PDB header:translation
Chain: B: PDB Molecule:elongation factor 1-beta;
PDBTitle: crystal structure of translation elongation factor ef-1 beta from2 pyrococcus horikoshii
|
| 34 | d3ceda1 |
|
not modelled |
22.6 |
13 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:NIL domain-like |
| 35 | c2zkre_ |
|
not modelled |
21.8 |
11 |
PDB header:ribosomal protein/rna
Chain: E: PDB Molecule:rna expansion segment es7 part ii;
PDBTitle: structure of a mammalian ribosomal 60s subunit within an2 80s complex obtained by docking homology models of the rna3 and proteins into an 8.7 a cryo-em map
|
| 36 | d1fx0a2 |
|
not modelled |
17.0 |
36 |
Fold:Domain of alpha and beta subunits of F1 ATP synthase-like
Superfamily:N-terminal domain of alpha and beta subunits of F1 ATP synthase
Family:N-terminal domain of alpha and beta subunits of F1 ATP synthase |
| 37 | d2jdia2 |
|
not modelled |
16.0 |
36 |
Fold:Domain of alpha and beta subunits of F1 ATP synthase-like
Superfamily:N-terminal domain of alpha and beta subunits of F1 ATP synthase
Family:N-terminal domain of alpha and beta subunits of F1 ATP synthase |
| 38 | c3gwpA_ |
|
not modelled |
15.9 |
19 |
PDB header:lyase
Chain: A: PDB Molecule:carbon-sulfur lyase involved in aluminum resistance;
PDBTitle: crystal structure of carbon-sulfur lyase involved in aluminum2 resistance (yp_878183.1) from clostridium novyi nt at 2.90 a3 resolution
|
| 39 | c3ucoB_ |
|
not modelled |
15.2 |
18 |
PDB header:lyase/lyase inhibitor
Chain: B: PDB Molecule:carbonic anhydrase;
PDBTitle: coccomyxa beta-carbonic anhydrase in complex with iodide
|
| 40 | c1jrjA_ |
|
not modelled |
14.3 |
53 |
PDB header:hormone/growth factor
Chain: A: PDB Molecule:exendin-4;
PDBTitle: solution structure of exendin-4 in 30-vol% trifluoroethanol
|
| 41 | c2dclB_ |
|
not modelled |
12.8 |
14 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:hypothetical upf0166 protein ph1503;
PDBTitle: structure of ph1503 protein from pyrococcus horikoshii ot3
|
| 42 | c2a5vB_ |
|
not modelled |
12.2 |
27 |
PDB header:lyase
Chain: B: PDB Molecule:carbonic anhydrase (carbonate dehydratase) (carbonic
PDBTitle: crystal structure of m. tuberculosis beta carbonic anhydrase, rv3588c,2 tetrameric form
|
| 43 | d2cz4a1 |
|
not modelled |
11.8 |
19 |
Fold:Ferredoxin-like
Superfamily:GlnB-like
Family:Prokaryotic signal transducing protein |
| 44 | d1d1ra_ |
|
not modelled |
11.7 |
16 |
Fold:eIF1-like
Superfamily:eIF1-like
Family:eIF1-like |
| 45 | c1d1rA_ |
|
not modelled |
11.7 |
16 |
PDB header:structural genomics
Chain: A: PDB Molecule:hypothetical 11.4 kd protein ycih in pyrf-osmb
PDBTitle: nmr solution structure of the product of the e. coli ycih2 gene.
|
| 46 | c2jz2A_ |
|
not modelled |
10.6 |
19 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:ssl0352 protein;
PDBTitle: solution nmr structure of ssl0352 protein from synechocystis sp. pcc2 6803. northeast structural genomics consortium target sgr42
|
| 47 | d1vqoh1 |
|
not modelled |
8.6 |
16 |
Fold:alpha/beta-Hammerhead
Superfamily:Ribosomal protein L16p/L10e
Family:Ribosomal protein L10e |
| 48 | c2qv6D_ |
|
not modelled |
8.3 |
10 |
PDB header:hydrolase
Chain: D: PDB Molecule:gtp cyclohydrolase iii;
PDBTitle: gtp cyclohydrolase iii from m. jannaschii (mj0145)2 complexed with gtp and metal ions
|
| 49 | d1maba2 |
|
not modelled |
7.8 |
36 |
Fold:Domain of alpha and beta subunits of F1 ATP synthase-like
Superfamily:N-terminal domain of alpha and beta subunits of F1 ATP synthase
Family:N-terminal domain of alpha and beta subunits of F1 ATP synthase |
| 50 | c3eyxB_ |
|
not modelled |
7.6 |
25 |
PDB header:lyase
Chain: B: PDB Molecule:carbonic anhydrase;
PDBTitle: crystal structure of carbonic anhydrase nce103 from2 saccharomyces cerevisiae
|
| 51 | d2fyxa1 |
|
not modelled |
7.5 |
15 |
Fold:Ferredoxin-like
Superfamily:Transposase IS200-like
Family:Transposase IS200-like |
| 52 | c3qm2A_ |
|
not modelled |
7.4 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:phosphoserine aminotransferase;
PDBTitle: 2.25 angstrom crystal structure of phosphoserine aminotransferase2 (serc) from salmonella enterica subsp. enterica serovar typhimurium
|
| 53 | c1iq8B_ |
|
not modelled |
7.2 |
13 |
PDB header:transferase
Chain: B: PDB Molecule:archaeosine trna-guanine transglycosylase;
PDBTitle: crystal structure of archaeosine trna-guanine2 transglycosylase from pyrococcus horikoshii
|
| 54 | d2vjva1 |
|
not modelled |
7.2 |
20 |
Fold:Ferredoxin-like
Superfamily:Transposase IS200-like
Family:Transposase IS200-like |
| 55 | c1vi7A_ |
|
not modelled |
6.9 |
9 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein yigz;
PDBTitle: crystal structure of an hypothetical protein
|
| 56 | d1nvma1 |
|
not modelled |
6.8 |
19 |
Fold:RuvA C-terminal domain-like
Superfamily:post-HMGL domain-like
Family:DmpG/LeuA communication domain-like |
| 57 | c1d0rA_ |
|
not modelled |
6.7 |
27 |
PDB header:hormone/growth factor
Chain: A: PDB Molecule:glucagon-like peptide-1-(7-36)-amide;
PDBTitle: solution structure of glucagon-like peptide-1-(7-36)-amide2 in trifluoroethanol/water
|
| 58 | c3c9gB_ |
|
not modelled |
6.6 |
3 |
PDB header:nucleotide binding protein
Chain: B: PDB Molecule:upf0200/upf0201 protein af_1395;
PDBTitle: crystal structure of uncharacterized upf0201 protein af_135
|
| 59 | d1ddza1 |
|
not modelled |
6.5 |
19 |
Fold:Resolvase-like
Superfamily:beta-carbonic anhydrase, cab
Family:beta-carbonic anhydrase, cab |
| 60 | c2zkrh_ |
|
not modelled |
6.4 |
16 |
PDB header:ribosomal protein/rna
Chain: H: PDB Molecule:rna expansion segment es12;
PDBTitle: structure of a mammalian ribosomal 60s subunit within an2 80s complex obtained by docking homology models of the rna3 and proteins into an 8.7 a cryo-em map
|
| 61 | d2zgwa1 |
|
not modelled |
6.3 |
47 |
Fold:SH3-like barrel
Superfamily:C-terminal domain of transcriptional repressors
Family:Biotin repressor (BirA) |
| 62 | c3lasA_ |
|
not modelled |
6.2 |
31 |
PDB header:lyase
Chain: A: PDB Molecule:putative carbonic anhydrase;
PDBTitle: crystal structure of carbonic anhydrase from streptococcus mutans to2 1.4 angstrom resolution
|
| 63 | d1whra_ |
|
not modelled |
5.9 |
14 |
Fold:IF3-like
Superfamily:R3H domain
Family:R3H domain |
| 64 | c2ldiA_ |
|
not modelled |
5.9 |
9 |
PDB header:hydrolase
Chain: A: PDB Molecule:zinc-transporting atpase;
PDBTitle: nmr solution structure of ziaan sub mutant
|
| 65 | c2qz4A_ |
|
not modelled |
5.8 |
18 |
PDB header:hydrolase
Chain: A: PDB Molecule:paraplegin;
PDBTitle: human paraplegin, aaa domain in complex with adp
|
| 66 | c2ju5A_ |
|
not modelled |
5.7 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:thioredoxin disulfide isomerase;
PDBTitle: dsbh oxidoreductase
|
| 67 | c2a8cE_ |
|
not modelled |
5.6 |
24 |
PDB header:lyase
Chain: E: PDB Molecule:carbonic anhydrase 2;
PDBTitle: haemophilus influenzae beta-carbonic anhydrase
|
| 68 | d1ddza2 |
|
not modelled |
5.5 |
20 |
Fold:Resolvase-like
Superfamily:beta-carbonic anhydrase, cab
Family:beta-carbonic anhydrase, cab |
| 69 | d2hh8a1 |
|
not modelled |
5.4 |
26 |
Fold:YdfO-like
Superfamily:YdfO-like
Family:YdfO-like |
| 70 | d1skyb2 |
|
not modelled |
5.3 |
33 |
Fold:Domain of alpha and beta subunits of F1 ATP synthase-like
Superfamily:N-terminal domain of alpha and beta subunits of F1 ATP synthase
Family:N-terminal domain of alpha and beta subunits of F1 ATP synthase |