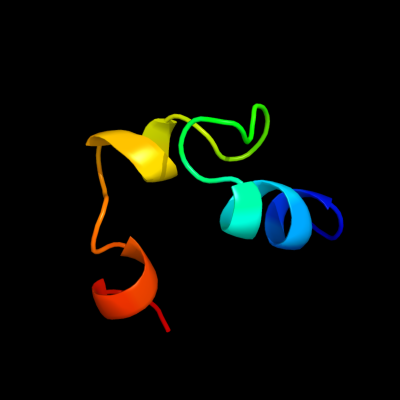
1 c2yukA_
38.4
22
PDB header: transferaseChain: A: PDB Molecule: myeloid/lymphoid or mixed-lineage leukemiaPDBTitle: solution structure of the hmg box of human myeloid/lymphoid2 or mixed-lineage leukemia protein 3 homolog
2 c2ph5A_
28.3
36
PDB header: transferaseChain: A: PDB Molecule: homospermidine synthase;PDBTitle: crystal structure of the homospermidine synthase hss from legionella2 pneumophila in complex with nad, northeast structural genomics target3 lgr54
3 d1lnla2
21.0
21
Fold: C-terminal domain of mollusc hemocyaninSuperfamily: C-terminal domain of mollusc hemocyaninFamily: C-terminal domain of mollusc hemocyanin4 d1qnta2
19.6
36
Fold: Ribonuclease H-like motifSuperfamily: Methylated DNA-protein cysteine methyltransferase domainFamily: Methylated DNA-protein cysteine methyltransferase domain5 d1iwpg_
15.6
18
Fold: Open three-helical up-and-down bundleSuperfamily: Diol dehydratase, gamma subunitFamily: Diol dehydratase, gamma subunit6 d1eexg_
15.6
21
Fold: Open three-helical up-and-down bundleSuperfamily: Diol dehydratase, gamma subunitFamily: Diol dehydratase, gamma subunit7 c1uc5M_
15.6
21
PDB header: lyaseChain: M: PDB Molecule: diol dehydrase gamma subunit;PDBTitle: structure of diol dehydratase complexed with (r)-1,2-2 propanediol
8 c2wbqA_
15.0
20
PDB header: oxidoreductaseChain: A: PDB Molecule: l-arginine beta-hydroxylase;PDBTitle: crystal structure of vioc in complex with (2s,3s)-2 hydroxyarginine
9 d2al1a1
14.9
24
Fold: TIM beta/alpha-barrelSuperfamily: Enolase C-terminal domain-likeFamily: Enolase10 d1k1fa_
14.8
43
Fold: Bcr-Abl oncoprotein oligomerization domainSuperfamily: Bcr-Abl oncoprotein oligomerization domainFamily: Bcr-Abl oncoprotein oligomerization domain11 c2blcA_
14.3
26
PDB header: oxidoreductaseChain: A: PDB Molecule: dihydrofolate reductase-thymidylate synthase;PDBTitle: sp21 double mutant p. vivax dihydrofolate reductase in2 complex with des-chloropyrimethamine
12 c2y94C_
14.3
13
PDB header: transferaseChain: C: PDB Molecule: 5'-amp-activated protein kinase catalytic subunit alpha-1;PDBTitle: structure of an active form of mammalian ampk
13 d1bg3a1
13.9
22
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: Hexokinase14 d1j3ka_
13.5
26
Fold: Dihydrofolate reductase-likeSuperfamily: Dihydrofolate reductase-likeFamily: Dihydrofolate reductases15 c3dg8B_
13.4
26
PDB header: oxidoreductase, transferaseChain: B: PDB Molecule: bifunctional dihydrofolate reductase-thymidylatePDBTitle: quadruple mutant (n51i+c59r+s108n+i164l) plasmodium2 falciparum dihydrofolate reductase-thymidylate synthase3 (pfdhfr-ts) complexed with rjf670, nadph, and dump
16 d1bdga1
12.9
15
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: Hexokinase17 d1js8a2
12.7
15
Fold: C-terminal domain of mollusc hemocyaninSuperfamily: C-terminal domain of mollusc hemocyaninFamily: C-terminal domain of mollusc hemocyanin18 c3lsgD_
12.5
14
PDB header: transcription regulatorChain: D: PDB Molecule: two-component response regulator yesn;PDBTitle: the crystal structure of the c-terminal domain of the two-2 component response regulator yesn from fusobacterium3 nucleatum subsp. nucleatum atcc 25586
19 c2r6fA_
12.3
18
PDB header: hydrolaseChain: A: PDB Molecule: excinuclease abc subunit a;PDBTitle: crystal structure of bacillus stearothermophilus uvra
20 c2l1nA_
11.4
44
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: solution nmr structure of the protein yp_399305.1
21 c2vbeA_
not modelled
10.8
16
PDB header: viral proteinChain: A: PDB Molecule: tailspike-protein;PDBTitle: tailspike protein of bacteriophage sf6
22 c3pmmA_
not modelled
10.3
29
PDB header: hydrolaseChain: A: PDB Molecule: putative cytoplasmic protein;PDBTitle: the crystal structure of a possible member of gh105 family from2 klebsiella pneumoniae subsp. pneumoniae mgh 78578
23 d1pdza1
not modelled
9.9
13
Fold: TIM beta/alpha-barrelSuperfamily: Enolase C-terminal domain-likeFamily: Enolase24 c2j4tB_
not modelled
9.7
27
PDB header: hydrolaseChain: B: PDB Molecule: angiogenin-4;PDBTitle: biological and structural features of murine angiogenin-4,2 an angiogenic protein
25 c2kjgA_
not modelled
9.6
14
PDB header: metal binding proteinChain: A: PDB Molecule: archaeal protein sso6904;PDBTitle: solution structure of an archaeal protein sso6904 from2 hyperthermophilic sulfolobus solfataricus
26 d1t44g_
not modelled
9.5
15
Fold: Gelsolin-likeSuperfamily: Actin depolymerizing proteinsFamily: Gelsolin-like27 c3cuoB_
not modelled
9.4
27
PDB header: transcription regulatorChain: B: PDB Molecule: uncharacterized hth-type transcriptional regulator ygav;PDBTitle: crystal structure of the predicted dna-binding transcriptional2 regulator from e. coli
28 c1skhA_
not modelled
9.1
50
PDB header: unknown functionChain: A: PDB Molecule: major prion protein 2;PDBTitle: n-terminal (1-30) of bovine prion protein
29 d1czan1
not modelled
9.1
21
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: Hexokinase30 c2vq8A_
not modelled
9.0
36
PDB header: hydrolaseChain: A: PDB Molecule: rnase zf-1a;PDBTitle: rnase zf-1a
31 c2lbbA_
not modelled
9.0
38
PDB header: protein bindingChain: A: PDB Molecule: acyl coa binding protein;PDBTitle: solution structure of acyl coa binding protein from babesia bovis t2bo
32 c3cw1A_
not modelled
8.8
24
PDB header: splicingChain: A: PDB Molecule: small nuclear ribonucleoprotein-associated proteins b andPDBTitle: crystal structure of human spliceosomal u1 snrnp
33 d1l6sa_
not modelled
8.8
26
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: 5-aminolaevulinate dehydratase, ALAD (porphobilinogen synthase)34 c3ixzB_
not modelled
8.8
8
PDB header: hydrolaseChain: B: PDB Molecule: potassium-transporting atpase subunit beta;PDBTitle: pig gastric h+/k+-atpase complexed with aluminium fluoride
35 c2w7nA_
not modelled
8.4
20
PDB header: transcription/dnaChain: A: PDB Molecule: trfb transcriptional repressor protein;PDBTitle: crystal structure of kora bound to operator dna: insight2 into repressor cooperation in rp4 gene regulation
36 d1bg3a3
not modelled
8.3
15
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: Hexokinase37 d1j72a1
not modelled
8.2
19
Fold: Gelsolin-likeSuperfamily: Actin depolymerizing proteinsFamily: Gelsolin-like38 d1dkua1
not modelled
8.1
26
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosylpyrophosphate synthetase-like39 c3ljeA_
not modelled
8.0
27
PDB header: hydrolaseChain: A: PDB Molecule: zebrafish rnase5;PDBTitle: the x-ray structure of zebrafish rnase5
40 d1gv7a_
not modelled
7.9
45
Fold: RNase A-likeSuperfamily: RNase A-likeFamily: Ribonuclease A-like41 c3l6wB_
not modelled
7.9
15
PDB header: oxygen bindingChain: B: PDB Molecule: hemocyanin 1;PDBTitle: structure of the collar functional unit (klh1-h) of keyhole2 limpet hemocyanin
42 d1m58a_
not modelled
7.9
20
Fold: RNase A-likeSuperfamily: RNase A-likeFamily: Ribonuclease A-like43 c2vq9A_
not modelled
7.8
27
PDB header: hydrolaseChain: A: PDB Molecule: rnase 1;PDBTitle: rnase zf-3e
44 d1oj8a_
not modelled
7.7
20
Fold: RNase A-likeSuperfamily: RNase A-likeFamily: Ribonuclease A-like45 c2p6zA_
not modelled
7.6
13
PDB header: hydrolaseChain: A: PDB Molecule: recombinant amphinase-2;PDBTitle: enzymatic and structural characterisation of amphinase, a2 novel cytotoxic ribonuclease from rana pipiens oocytes
46 d1m07a_
not modelled
7.6
13
Fold: RNase A-likeSuperfamily: RNase A-likeFamily: Ribonuclease A-like47 d1m5ha1
not modelled
7.5
35
Fold: Ferredoxin-likeSuperfamily: Formylmethanofuran:tetrahydromethanopterin formyltransferaseFamily: Formylmethanofuran:tetrahydromethanopterin formyltransferase48 d1e0na_
not modelled
7.5
83
Fold: WW domain-likeSuperfamily: WW domainFamily: WW domain49 d1onca_
not modelled
7.5
13
Fold: RNase A-likeSuperfamily: RNase A-likeFamily: Ribonuclease A-like50 d1czan3
not modelled
7.4
15
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: Hexokinase51 d1d0na4
not modelled
7.2
14
Fold: Gelsolin-likeSuperfamily: Actin depolymerizing proteinsFamily: Gelsolin-like52 c2jpiA_
not modelled
7.2
17
PDB header: structural genomicsChain: A: PDB Molecule: hypothetical protein;PDBTitle: chemical shift assignments of pa4090 from pseudomonas2 aeruginosa
53 d1k59a_
not modelled
7.1
45
Fold: RNase A-likeSuperfamily: RNase A-likeFamily: Ribonuclease A-like54 c3pgwB_
not modelled
7.1
24
PDB header: splicing/dna/rnaChain: B: PDB Molecule: sm b;PDBTitle: crystal structure of human u1 snrnp
55 d3buxb1
not modelled
7.0
12
Fold: EF Hand-likeSuperfamily: EF-handFamily: EF-hand modules in multidomain proteins56 d1un3a_
not modelled
6.9
45
Fold: RNase A-likeSuperfamily: RNase A-likeFamily: Ribonuclease A-like57 d1v4sa1
not modelled
6.9
15
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: Hexokinase58 d1agia_
not modelled
6.9
36
Fold: RNase A-likeSuperfamily: RNase A-likeFamily: Ribonuclease A-like59 d2akza1
not modelled
6.8
13
Fold: TIM beta/alpha-barrelSuperfamily: Enolase C-terminal domain-likeFamily: Enolase60 c3obkH_
not modelled
6.7
30
PDB header: lyaseChain: H: PDB Molecule: delta-aminolevulinic acid dehydratase;PDBTitle: crystal structure of delta-aminolevulinic acid dehydratase2 (porphobilinogen synthase) from toxoplasma gondii me49 in complex3 with the reaction product porphobilinogen
61 d1kvza_
not modelled
6.7
20
Fold: RNase A-likeSuperfamily: RNase A-likeFamily: Ribonuclease A-like62 d2fug71
not modelled
6.5
24
Fold: N domain of copper amine oxidase-likeSuperfamily: Frataxin/Nqo15-likeFamily: Nqo15-like63 c2zpoA_
not modelled
6.3
36
PDB header: hydrolaseChain: A: PDB Molecule: ribonuclease;PDBTitle: crystal structure of green turtle egg white ribonuclease
64 c3oioA_
not modelled
6.1
16
PDB header: transcription regulatorChain: A: PDB Molecule: transcriptional regulator (arac-type dna-binding domain-PDBTitle: crystal structure of transcriptional regulator (arac-type dna-binding2 domain-containing proteins) from chromobacterium violaceum
65 c2vefB_
not modelled
5.9
20
PDB header: transferaseChain: B: PDB Molecule: dihydropteroate synthase;PDBTitle: dihydropteroate synthase from streptococcus pneumoniae
66 d1nrja_
not modelled
5.9
25
Fold: Profilin-likeSuperfamily: SNARE-likeFamily: SRP alpha N-terminal domain-like67 c2k9sA_
not modelled
5.8
13
PDB header: transcriptionChain: A: PDB Molecule: arabinose operon regulatory protein;PDBTitle: solution structure of dna binding domain of e. coli arac
68 c3mklB_
not modelled
5.7
17
PDB header: transcription regulatorChain: B: PDB Molecule: hth-type transcriptional regulator gadx;PDBTitle: crystal structure of dna-binding transcriptional dual regulator from2 escherichia coli k-12
69 d1iyxa1
not modelled
5.7
15
Fold: TIM beta/alpha-barrelSuperfamily: Enolase C-terminal domain-likeFamily: Enolase70 d2hkja2
not modelled
5.6
21
Fold: Ribosomal protein S5 domain 2-likeSuperfamily: Ribosomal protein S5 domain 2-likeFamily: DNA gyrase/MutL, second domain71 d2ebfx3
not modelled
5.6
36
Fold: Cysteine proteinasesSuperfamily: Cysteine proteinasesFamily: PMT C-terminal domain like72 d1rkea1
not modelled
5.6
25
Fold: Four-helical up-and-down bundleSuperfamily: alpha-catenin/vinculin-likeFamily: alpha-catenin/vinculin73 d5ruba1
not modelled
5.6
23
Fold: TIM beta/alpha-barrelSuperfamily: RuBisCo, C-terminal domainFamily: RuBisCo, large subunit, C-terminal domain74 d1w6ta1
not modelled
5.5
19
Fold: TIM beta/alpha-barrelSuperfamily: Enolase C-terminal domain-likeFamily: Enolase75 c3n9tA_
not modelled
5.5
18
PDB header: oxidoreductaseChain: A: PDB Molecule: pnpc;PDBTitle: cryatal structure of hydroxyquinol 1,2-dioxygenase from pseudomonas2 putida dll-e4
76 d1qo7a_
not modelled
5.3
15
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Epoxide hydrolase77 d1hbka_
not modelled
5.2
23
Fold: Acyl-CoA binding protein-likeSuperfamily: Acyl-CoA binding proteinFamily: Acyl-CoA binding protein78 c2copA_
not modelled
5.1
25
PDB header: lipid binding proteinChain: A: PDB Molecule: acyl-coenzyme a binding domain containing 6;PDBTitle: solution structure of rsgi ruh-040, an acbp domain from2 human cdna
79 c2k29A_
not modelled
5.1
29
PDB header: transcriptionChain: A: PDB Molecule: antitoxin relb;PDBTitle: structure of the dbd domain of e. coli antitoxin relb
80 d2qn6b1
not modelled
5.1
16
Fold: Ferredoxin-likeSuperfamily: eIF-2-alpha, C-terminal domainFamily: eIF-2-alpha, C-terminal domain81 c2mltA_
not modelled
5.0
32
PDB header: toxin (hemolytic polypeptide)Chain: A: PDB Molecule: melittin;PDBTitle: melittin