| 1 |
|
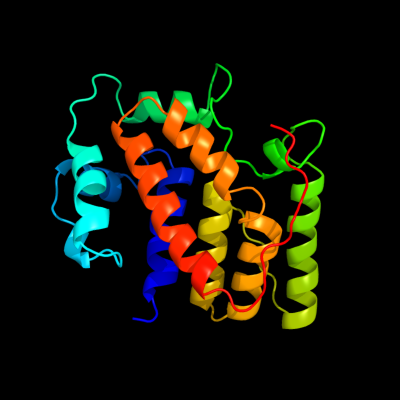
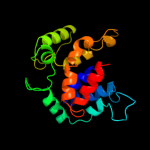
PDB 1s9u chain A
Region: 6 - 204
Aligned: 193
Modelled: 199
Confidence: 100.0%
Identity: 76%
Fold: TorD-like
Superfamily: TorD-like
Family: TorD-like
Phyre2
| 2 |
|
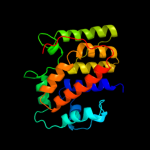
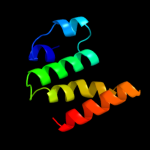
PDB 1n1c chain A
Region: 4 - 194
Aligned: 185
Modelled: 191
Confidence: 100.0%
Identity: 17%
Fold: TorD-like
Superfamily: TorD-like
Family: TorD-like
Phyre2
| 3 |
|
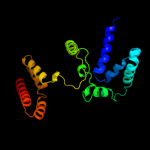
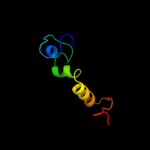
PDB 2o9x chain A
Region: 7 - 187
Aligned: 154
Modelled: 179
Confidence: 100.0%
Identity: 20%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:reductase, assembly protein;
PDBTitle: crystal structure of a putative redox enzyme maturation protein from2 archaeoglobus fulgidus
Phyre2
| 4 |
|
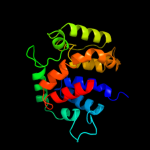
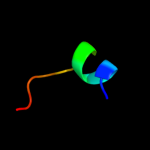
PDB 2o9x chain A domain 1
Region: 13 - 188
Aligned: 149
Modelled: 174
Confidence: 100.0%
Identity: 21%
Fold: TorD-like
Superfamily: TorD-like
Family: TorD-like
Phyre2
| 5 |
|
PDB 2idg chain A domain 1
Region: 106 - 182
Aligned: 74
Modelled: 76
Confidence: 99.7%
Identity: 19%
Fold: TorD-like
Superfamily: TorD-like
Family: TorD-like
Phyre2
| 6 |
|
PDB 1q8h chain A
Region: 117 - 131
Aligned: 15
Modelled: 15
Confidence: 17.2%
Identity: 27%
PDB header:metal binding protein
Chain: A: PDB Molecule:osteocalcin;
PDBTitle: crystal structure of porcine osteocalcin
Phyre2
| 7 |
|
PDB 1q8h chain A
Region: 117 - 131
Aligned: 15
Modelled: 15
Confidence: 17.2%
Identity: 27%
Fold: GLA-domain
Superfamily: GLA-domain
Family: GLA-domain
Phyre2
| 8 |
|
PDB 2r7m chain A
Region: 70 - 126
Aligned: 53
Modelled: 57
Confidence: 15.3%
Identity: 21%
PDB header:ligase
Chain: A: PDB Molecule:5-formaminoimidazole-4-carboxamide-1-(beta)-d-
PDBTitle: crystal structure of faicar synthetase (purp) from m.2 jannaschii complexed with amp
Phyre2
| 9 |
|
PDB 1m3v chain A domain 2
Region: 69 - 81
Aligned: 13
Modelled: 13
Confidence: 12.2%
Identity: 31%
Fold: Glucocorticoid receptor-like (DNA-binding domain)
Superfamily: Glucocorticoid receptor-like (DNA-binding domain)
Family: LIM domain
Phyre2
| 10 |
|
PDB 1q3m chain A
Region: 117 - 130
Aligned: 14
Modelled: 14
Confidence: 12.1%
Identity: 29%
Fold: GLA-domain
Superfamily: GLA-domain
Family: GLA-domain
Phyre2
| 11 |
|
PDB 1ysg chain A domain 1
Region: 127 - 161
Aligned: 35
Modelled: 35
Confidence: 10.8%
Identity: 20%
Fold: Toxins' membrane translocation domains
Superfamily: Bcl-2 inhibitors of programmed cell death
Family: Bcl-2 inhibitors of programmed cell death
Phyre2
| 12 |
|
PDB 1yt3 chain A domain 2
Region: 171 - 197
Aligned: 27
Modelled: 27
Confidence: 8.8%
Identity: 15%
Fold: SAM domain-like
Superfamily: HRDC-like
Family: RNase D C-terminal domains
Phyre2
| 13 |
|
PDB 1lcd chain A
Region: 89 - 119
Aligned: 31
Modelled: 31
Confidence: 8.4%
Identity: 13%
Fold: lambda repressor-like DNA-binding domains
Superfamily: lambda repressor-like DNA-binding domains
Family: GalR/LacI-like bacterial regulator
Phyre2
| 14 |
|
PDB 2qec chain A
Region: 105 - 127
Aligned: 23
Modelled: 23
Confidence: 7.3%
Identity: 13%
PDB header:transferase
Chain: A: PDB Molecule:histone acetyltransferase hpa2 and related
PDBTitle: crystal structure of histone acetyltransferase hpa2 and related2 acetyltransferase (np_600742.1) from corynebacterium glutamicum atcc3 13032 at 1.90 a resolution
Phyre2
| 15 |
|
PDB 3qbr chain A
Region: 127 - 159
Aligned: 33
Modelled: 33
Confidence: 7.2%
Identity: 12%
PDB header:apoptosis
Chain: A: PDB Molecule:sjchgc06286 protein;
PDBTitle: bakbh3 in complex with sja
Phyre2
| 16 |
|
PDB 2pon chain B domain 1
Region: 127 - 161
Aligned: 35
Modelled: 35
Confidence: 6.9%
Identity: 20%
Fold: Toxins' membrane translocation domains
Superfamily: Bcl-2 inhibitors of programmed cell death
Family: Bcl-2 inhibitors of programmed cell death
Phyre2
| 17 |
|
PDB 1g5m chain A
Region: 128 - 161
Aligned: 34
Modelled: 34
Confidence: 6.5%
Identity: 15%
Fold: Toxins' membrane translocation domains
Superfamily: Bcl-2 inhibitors of programmed cell death
Family: Bcl-2 inhibitors of programmed cell death
Phyre2
| 18 |
|
PDB 1pq1 chain A
Region: 127 - 161
Aligned: 35
Modelled: 35
Confidence: 6.0%
Identity: 20%
Fold: Toxins' membrane translocation domains
Superfamily: Bcl-2 inhibitors of programmed cell death
Family: Bcl-2 inhibitors of programmed cell death
Phyre2
| 19 |
|
PDB 1qpz chain A domain 1
Region: 89 - 118
Aligned: 30
Modelled: 30
Confidence: 5.9%
Identity: 10%
Fold: lambda repressor-like DNA-binding domains
Superfamily: lambda repressor-like DNA-binding domains
Family: GalR/LacI-like bacterial regulator
Phyre2
| 20 |
|
PDB 1f16 chain A
Region: 127 - 161
Aligned: 35
Modelled: 35
Confidence: 5.7%
Identity: 20%
Fold: Toxins' membrane translocation domains
Superfamily: Bcl-2 inhibitors of programmed cell death
Family: Bcl-2 inhibitors of programmed cell death
Phyre2
| 21 |
|