| 1 |
|
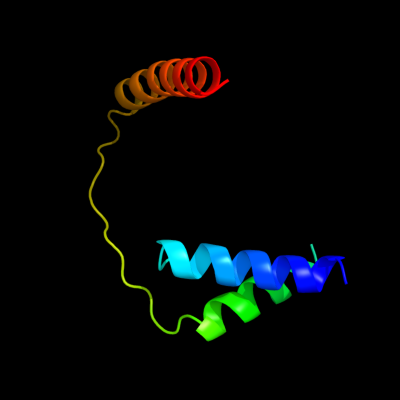
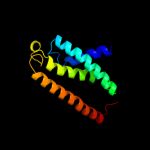
PDB 3org chain B
Region: 12 - 82
Aligned: 69
Modelled: 71
Confidence: 41.0%
Identity: 26%
PDB header:transport protein
Chain: B: PDB Molecule:cmclc;
PDBTitle: crystal structure of a eukaryotic clc transporter
Phyre2
| 2 |
|
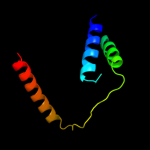
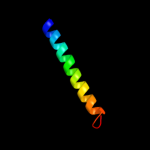
PDB 3hd6 chain A
Region: 7 - 156
Aligned: 146
Modelled: 150
Confidence: 18.5%
Identity: 9%
PDB header:membrane protein, transport protein
Chain: A: PDB Molecule:ammonium transporter rh type c;
PDBTitle: crystal structure of the human rhesus glycoprotein rhcg
Phyre2
| 3 |
|
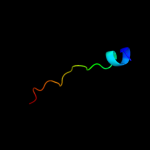
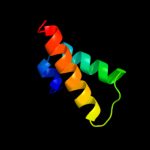
PDB 1cii chain A
Region: 16 - 48
Aligned: 33
Modelled: 33
Confidence: 9.4%
Identity: 15%
PDB header:transmembrane protein
Chain: A: PDB Molecule:colicin ia;
PDBTitle: colicin ia
Phyre2
| 4 |
|
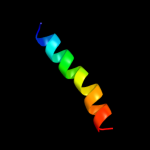
PDB 2uwj chain F
Region: 100 - 112
Aligned: 13
Modelled: 13
Confidence: 9.0%
Identity: 8%
PDB header:chaperone
Chain: F: PDB Molecule:type iii export protein pscf;
PDBTitle: structure of the heterotrimeric complex which regulates2 type iii secretion needle formation
Phyre2
| 5 |
|
PDB 2k1a chain A
Region: 135 - 155
Aligned: 21
Modelled: 21
Confidence: 8.2%
Identity: 33%
PDB header:cell adhesion
Chain: A: PDB Molecule:integrin alpha-iib;
PDBTitle: bicelle-embedded integrin alpha(iib) transmembrane segment
Phyre2
| 6 |
|
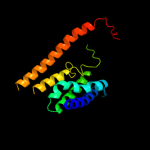
PDB 2b2h chain A
Region: 8 - 157
Aligned: 126
Modelled: 126
Confidence: 7.7%
Identity: 12%
PDB header:transport protein
Chain: A: PDB Molecule:ammonium transporter;
PDBTitle: ammonium transporter amt-1 from a. fulgidus (as)
Phyre2
| 7 |
|
PDB 2jwa chain A
Region: 125 - 156
Aligned: 32
Modelled: 32
Confidence: 6.9%
Identity: 16%
PDB header:transferase
Chain: A: PDB Molecule:receptor tyrosine-protein kinase erbb-2;
PDBTitle: erbb2 transmembrane segment dimer spatial structure
Phyre2
| 8 |
|
PDB 1tlq chain A
Region: 35 - 80
Aligned: 46
Modelled: 46
Confidence: 6.1%
Identity: 17%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein ypjq;
PDBTitle: crystal structure of protein ypjq from bacillus subtilis, pfam duf64
Phyre2
| 9 |
|
PDB 1tlq chain A
Region: 35 - 80
Aligned: 46
Modelled: 46
Confidence: 6.1%
Identity: 17%
Fold: YutG-like
Superfamily: YutG-like
Family: YutG-like
Phyre2
| 10 |
|
PDB 1jb0 chain M
Region: 83 - 104
Aligned: 22
Modelled: 22
Confidence: 6.0%
Identity: 27%
Fold: Single transmembrane helix
Superfamily: Subunit XII of photosystem I reaction centre, PsaM
Family: Subunit XII of photosystem I reaction centre, PsaM
Phyre2