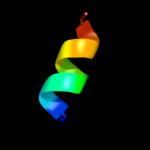| 1 |
|
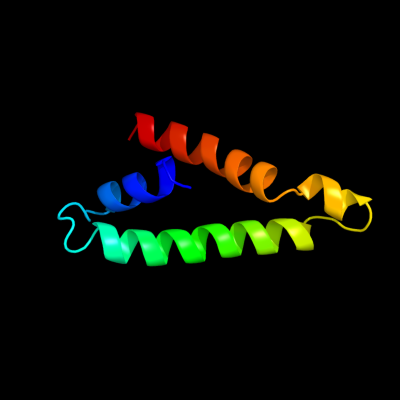
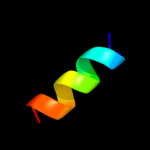
PDB 1l7v chain A
Region: 9 - 79
Aligned: 68
Modelled: 71
Confidence: 92.7%
Identity: 16%
Fold: ABC transporter involved in vitamin B12 uptake, BtuC
Superfamily: ABC transporter involved in vitamin B12 uptake, BtuC
Family: ABC transporter involved in vitamin B12 uptake, BtuC
Phyre2
| 2 |
|
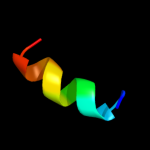
PDB 2nq2 chain A
Region: 9 - 80
Aligned: 68
Modelled: 72
Confidence: 88.7%
Identity: 19%
PDB header:metal transport
Chain: A: PDB Molecule:hypothetical abc transporter permease protein
PDBTitle: an inward-facing conformation of a putative metal-chelate2 type abc transporter.
Phyre2
| 3 |
|
PDB 2ap8 chain A
Region: 34 - 46
Aligned: 13
Modelled: 13
Confidence: 19.3%
Identity: 46%
PDB header:antibiotic
Chain: A: PDB Molecule:bombinin h4;
PDBTitle: solution structure of bombinin h4 in dpc micelles
Phyre2
| 4 |
|
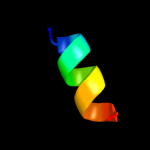
PDB 2ap7 chain A
Region: 34 - 46
Aligned: 13
Modelled: 13
Confidence: 19.1%
Identity: 46%
PDB header:antibiotic
Chain: A: PDB Molecule:bombinin h2;
PDBTitle: solution structure of bombinin h2 in dpc micelles
Phyre2
| 5 |
|
PDB 1xoo chain A
Region: 8 - 18
Aligned: 11
Modelled: 11
Confidence: 12.0%
Identity: 45%
PDB header:viral protein
Chain: A: PDB Molecule:hemagglutinin;
PDBTitle: nmr structure of g1s mutant of influenza hemagglutinin2 fusion peptide in dpc micelles at ph 5
Phyre2
| 6 |
|
PDB 2l4g chain A
Region: 8 - 19
Aligned: 12
Modelled: 12
Confidence: 11.6%
Identity: 42%
PDB header:viral protein
Chain: A: PDB Molecule:haemagglutinin;
PDBTitle: influenza haemagglutinin fusion peptide mutant g13a
Phyre2
| 7 |
|
PDB 2kxa chain A
Region: 8 - 16
Aligned: 9
Modelled: 9
Confidence: 11.2%
Identity: 56%
PDB header:viral protein, immune system
Chain: A: PDB Molecule:haemagglutinin ha2 chain peptide;
PDBTitle: the hemagglutinin fusion peptide (h1 subtype) at ph 7.4
Phyre2
| 8 |
|
PDB 1ibo chain A
Region: 8 - 18
Aligned: 11
Modelled: 11
Confidence: 11.0%
Identity: 45%
PDB header:viral protein
Chain: A: PDB Molecule:hemagglutinin ha2 chain peptide;
PDBTitle: nmr structure of hemagglutinin fusion peptide in dpc2 micelles at ph 7.4
Phyre2
| 9 |
|
PDB 1ibn chain A
Region: 8 - 18
Aligned: 11
Modelled: 11
Confidence: 11.0%
Identity: 45%
PDB header:viral protein
Chain: A: PDB Molecule:hemagglutinin ha2 chain peptide;
PDBTitle: nmr structure of hemagglutinin fusion peptide in dpc2 micelles at ph 5
Phyre2
| 10 |
|
PDB 1xop chain A
Region: 8 - 18
Aligned: 11
Modelled: 11
Confidence: 10.9%
Identity: 45%
PDB header:viral protein
Chain: A: PDB Molecule:hemagglutinin;
PDBTitle: nmr structure of g1v mutant of influenza hemagglutinin2 fusion peptide in dpc micelles at ph 5
Phyre2
| 11 |
|
PDB 2jrd chain A
Region: 8 - 18
Aligned: 11
Modelled: 11
Confidence: 7.0%
Identity: 45%
PDB header:viral protein
Chain: A: PDB Molecule:hemagglutinin;
PDBTitle: influenza hemagglutinin fusion domain mutant f9a
Phyre2
| 12 |
|
PDB 3c1i chain A
Region: 1 - 82
Aligned: 82
Modelled: 82
Confidence: 6.5%
Identity: 23%
PDB header:transport protein
Chain: A: PDB Molecule:ammonia channel;
PDBTitle: substrate binding, deprotonation and selectivity at the2 periplasmic entrance of the e. coli ammonia channel amtb
Phyre2
| 13 |
|
PDB 2kpe chain A
Region: 4 - 21
Aligned: 18
Modelled: 18
Confidence: 6.3%
Identity: 33%
PDB header:membrane protein
Chain: A: PDB Molecule:glycophorin-a;
PDBTitle: refined structure of glycophorin a transmembrane segment dimer in dpc2 micelles
Phyre2
| 14 |
|
PDB 2kpe chain B
Region: 4 - 21
Aligned: 18
Modelled: 18
Confidence: 6.3%
Identity: 33%
PDB header:membrane protein
Chain: B: PDB Molecule:glycophorin-a;
PDBTitle: refined structure of glycophorin a transmembrane segment dimer in dpc2 micelles
Phyre2