| 1 |
|
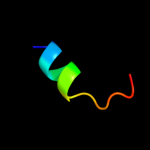
PDB 1kft chain A
Region: 3 - 44
Aligned: 39
Modelled: 42
Confidence: 16.3%
Identity: 33%
Fold: SAM domain-like
Superfamily: RuvA domain 2-like
Family: Excinuclease UvrC C-terminal domain
Phyre2
| 2 |
|
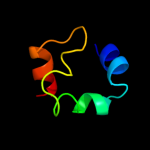
PDB 1kft chain A
Region: 3 - 44
Aligned: 39
Modelled: 42
Confidence: 16.3%
Identity: 33%
PDB header:dna binding protein
Chain: A: PDB Molecule:excinuclease abc subunit c;
PDBTitle: solution structure of the c-terminal domain of uvrc from e-2 coli
Phyre2
| 3 |
|
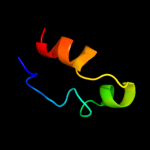
PDB 1qgi chain A
Region: 11 - 75
Aligned: 65
Modelled: 65
Confidence: 14.9%
Identity: 20%
Fold: Lysozyme-like
Superfamily: Lysozyme-like
Family: Chitosanase
Phyre2
| 4 |
|
PDB 1x2i chain A domain 1
Region: 3 - 44
Aligned: 39
Modelled: 42
Confidence: 13.9%
Identity: 26%
Fold: SAM domain-like
Superfamily: RuvA domain 2-like
Family: Hef domain-like
Phyre2
| 5 |
|
PDB 1pcf chain A
Region: 1 - 28
Aligned: 28
Modelled: 28
Confidence: 9.0%
Identity: 18%
Fold: ssDNA-binding transcriptional regulator domain
Superfamily: ssDNA-binding transcriptional regulator domain
Family: Transcriptional coactivator PC4 C-terminal domain
Phyre2
| 6 |
|
PDB 2aq0 chain A domain 1
Region: 9 - 44
Aligned: 32
Modelled: 36
Confidence: 8.3%
Identity: 25%
Fold: SAM domain-like
Superfamily: RuvA domain 2-like
Family: Hef domain-like
Phyre2
| 7 |
|
PDB 2i9c chain A domain 1
Region: 21 - 47
Aligned: 27
Modelled: 27
Confidence: 6.6%
Identity: 22%
Fold: alpha-alpha superhelix
Superfamily: ARM repeat
Family: RPA1889-like
Phyre2
| 8 |
|
PDB 2f22 chain A domain 1
Region: 47 - 70
Aligned: 24
Modelled: 22
Confidence: 6.2%
Identity: 38%
Fold: DinB/YfiT-like putative metalloenzymes
Superfamily: DinB/YfiT-like putative metalloenzymes
Family: DinB-like
Phyre2
| 9 |
|
PDB 2jtt chain D
Region: 38 - 50
Aligned: 13
Modelled: 13
Confidence: 5.8%
Identity: 38%
PDB header:calcium binding protein/antitumor protei
Chain: D: PDB Molecule:calcyclin-binding protein;
PDBTitle: solution structure of calcium loaded s100a6 bound to c-2 terminal siah-1 interacting protein
Phyre2
| 10 |
|
PDB 2bgw chain A domain 1
Region: 3 - 44
Aligned: 39
Modelled: 42
Confidence: 5.7%
Identity: 33%
Fold: SAM domain-like
Superfamily: RuvA domain 2-like
Family: Hef domain-like
Phyre2
| 11 |
|
PDB 1b22 chain A
Region: 15 - 45
Aligned: 28
Modelled: 31
Confidence: 5.2%
Identity: 21%
Fold: SAM domain-like
Superfamily: Rad51 N-terminal domain-like
Family: DNA repair protein Rad51, N-terminal domain
Phyre2