| 1 |
|
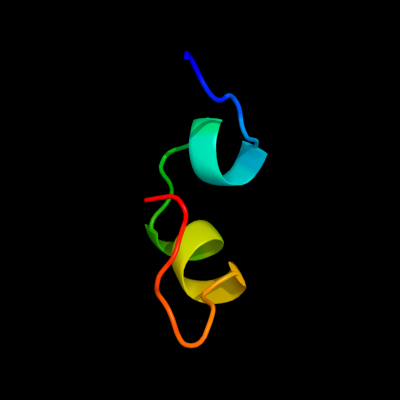
PDB 2z1d chain A
Region: 36 - 62
Aligned: 20
Modelled: 27
Confidence: 11.9%
Identity: 30%
PDB header:metal binding protein
Chain: A: PDB Molecule:hydrogenase expression/formation protein hypd;
PDBTitle: crystal structure of [nife] hydrogenase maturation protein, hypd from2 thermococcus kodakaraensis
Phyre2
| 2 |
|
PDB 1fjr chain A
Region: 36 - 69
Aligned: 34
Modelled: 34
Confidence: 8.0%
Identity: 24%
Fold: Methuselah ectodomain
Superfamily: Methuselah ectodomain
Family: Methuselah ectodomain
Phyre2
| 3 |
|
PDB 1icl chain A
Region: 46 - 60
Aligned: 15
Modelled: 15
Confidence: 6.0%
Identity: 47%
PDB header:de novo protein
Chain: A: PDB Molecule:th1ox;
PDBTitle: solution structure of designed beta-sheet mini-protein th1ox
Phyre2
| 4 |
|
PDB 3arc chain T
Region: 19 - 25
Aligned: 7
Modelled: 7
Confidence: 5.0%
Identity: 86%
PDB header:electron transport, photosynthesis
Chain: T: PDB Molecule:photosystem ii reaction center protein t;
PDBTitle: crystal structure of oxygen-evolving photosystem ii at 1.9 angstrom2 resolution
Phyre2
| 5 |
|
PDB 2axt chain T
Region: 19 - 25
Aligned: 7
Modelled: 7
Confidence: 5.0%
Identity: 86%
PDB header:electron transport
Chain: T: PDB Molecule:photosystem ii reaction center t protein;
PDBTitle: crystal structure of photosystem ii from thermosynechococcus elongatus
Phyre2
| 6 |
|
PDB 2axt chain T
Region: 19 - 25
Aligned: 7
Modelled: 7
Confidence: 5.0%
Identity: 86%
PDB header:electron transport
Chain: T: PDB Molecule:photosystem ii reaction center t protein;
PDBTitle: crystal structure of photosystem ii from thermosynechococcus elongatus
Phyre2
| 7 |
|
PDB 3a0b chain T
Region: 19 - 25
Aligned: 7
Modelled: 7
Confidence: 5.0%
Identity: 86%
PDB header:electron transport
Chain: T: PDB Molecule:photosystem ii reaction center protein t;
PDBTitle: crystal structure of br-substituted photosystem ii complex
Phyre2
| 8 |
|
PDB 2axt chain T domain 1
Region: 19 - 25
Aligned: 7
Modelled: 7
Confidence: 5.0%
Identity: 86%
Fold: Single transmembrane helix
Superfamily: Photosystem II reaction center protein T, PsbT
Family: PsbT-like
Phyre2
| 9 |
|
PDB 3a0h chain T
Region: 19 - 25
Aligned: 7
Modelled: 7
Confidence: 5.0%
Identity: 86%
PDB header:electron transport
Chain: T: PDB Molecule:photosystem ii reaction center protein t;
PDBTitle: crystal structure of i-substituted photosystem ii complex
Phyre2
| 10 |
|
PDB 3kzi chain T
Region: 19 - 25
Aligned: 7
Modelled: 7
Confidence: 5.0%
Identity: 86%
PDB header:electron transport
Chain: T: PDB Molecule:photosystem ii reaction center protein t;
PDBTitle: crystal structure of monomeric form of cyanobacterial photosystem ii
Phyre2