| 1 |
|
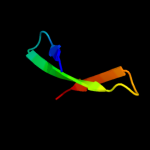
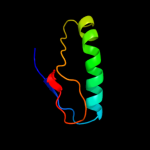
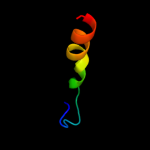
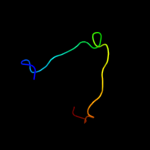
PDB 1d8j chain A
Region: 29 - 43
Aligned: 14
Modelled: 15
Confidence: 28.7%
Identity: 29%
Fold: DNA/RNA-binding 3-helical bundle
Superfamily: "Winged helix" DNA-binding domain
Family: The central core domain of TFIIE beta
Phyre2
| 2 |
|
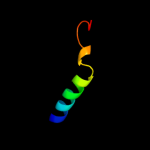
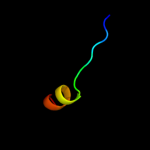
PDB 1z1b chain A domain 1
Region: 46 - 67
Aligned: 22
Modelled: 22
Confidence: 20.7%
Identity: 23%
Fold: DNA-binding domain
Superfamily: DNA-binding domain
Family: lambda integrase N-terminal domain
Phyre2
| 3 |
|
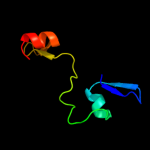
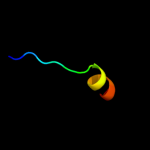
PDB 1kjk chain A
Region: 46 - 67
Aligned: 22
Modelled: 22
Confidence: 20.7%
Identity: 23%
PDB header:viral protein
Chain: A: PDB Molecule:integrase;
PDBTitle: solution structure of the lambda integrase amino-terminal2 domain
Phyre2
| 4 |
|
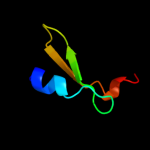
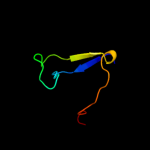
PDB 2jna chain A domain 1
Region: 2 - 31
Aligned: 27
Modelled: 30
Confidence: 16.8%
Identity: 56%
Fold: Dodecin subunit-like
Superfamily: YdgH-like
Family: YdgH-like
Phyre2
| 5 |
|
PDB 3ctz chain A
Region: 70 - 109
Aligned: 40
Modelled: 40
Confidence: 15.8%
Identity: 10%
PDB header:hydrolase
Chain: A: PDB Molecule:xaa-pro aminopeptidase 1;
PDBTitle: structure of human cytosolic x-prolyl aminopeptidase
Phyre2
| 6 |
|
PDB 2axt chain I domain 1
Region: 6 - 30
Aligned: 25
Modelled: 25
Confidence: 8.9%
Identity: 16%
Fold: Single transmembrane helix
Superfamily: Photosystem II reaction center protein I, PsbI
Family: PsbI-like
Phyre2
| 7 |
|
PDB 1lm7 chain A
Region: 55 - 110
Aligned: 55
Modelled: 56
Confidence: 8.8%
Identity: 16%
Fold: beta-hairpin-alpha-hairpin repeat
Superfamily: Plakin repeat
Family: Plakin repeat
Phyre2
| 8 |
|
PDB 1icc chain C
Region: 39 - 73
Aligned: 32
Modelled: 35
Confidence: 7.9%
Identity: 22%
Fold: Cytochrome b5-like heme/steroid binding domain
Superfamily: Cytochrome b5-like heme/steroid binding domain
Family: Cytochrome b5
Phyre2
| 9 |
|
PDB 1vjq chain B
Region: 52 - 102
Aligned: 43
Modelled: 51
Confidence: 6.6%
Identity: 19%
PDB header:structural genomics, de novo protein
Chain: B: PDB Molecule:designed protein;
PDBTitle: designed protein based on backbone conformation of2 procarboxypeptidase-a (1aye) with sidechains chosen for maximal3 predicted stability.
Phyre2
| 10 |
|
PDB 1s21 chain A
Region: 94 - 109
Aligned: 16
Modelled: 15
Confidence: 6.3%
Identity: 19%
Fold: ADP-ribosylation
Superfamily: ADP-ribosylation
Family: AvrPphF ORF2, a type III effector
Phyre2
| 11 |
|
PDB 1s21 chain A
Region: 94 - 109
Aligned: 16
Modelled: 15
Confidence: 6.3%
Identity: 19%
PDB header:chaperone
Chain: A: PDB Molecule:orf2;
PDBTitle: crystal structure of avrpphf orf2, a type iii effector from2 p. syringae
Phyre2
| 12 |
|
PDB 2rm4 chain A
Region: 22 - 62
Aligned: 41
Modelled: 41
Confidence: 6.0%
Identity: 22%
PDB header:protein binding
Chain: A: PDB Molecule:cg6311-pb;
PDBTitle: solution structure of the lsm domain of dm edc3 (enhancer2 of decapping 3)
Phyre2
| 13 |
|
PDB 1st6 chain A domain 4
Region: 89 - 112
Aligned: 24
Modelled: 24
Confidence: 5.9%
Identity: 25%
Fold: Four-helical up-and-down bundle
Superfamily: alpha-catenin/vinculin-like
Family: alpha-catenin/vinculin
Phyre2
| 14 |
|
PDB 2vc8 chain A
Region: 38 - 64
Aligned: 27
Modelled: 27
Confidence: 5.8%
Identity: 15%
PDB header:protein-binding
Chain: A: PDB Molecule:enhancer of mrna-decapping protein 3;
PDBTitle: crystal structure of the lsm domain of human edc3 (enhancer2 of decapping 3)
Phyre2