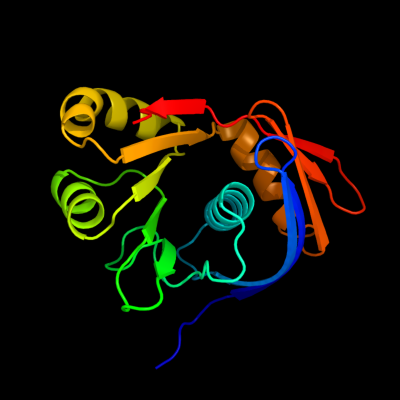
| 1 | d2g3wa1
|
|
 |
100.0 |
38 |
Fold:Restriction endonuclease-like
Superfamily:Restriction endonuclease-like
Family:YaeQ-like |
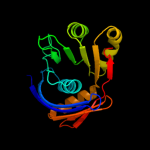
| 2 | c3c0uA_
|
|
 |
100.0 |
98 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein yaeq;
PDBTitle: crystal structure of e.coli yaeq protein
|
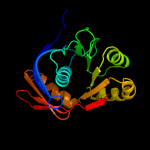
| 3 | d2ot9a1
|
|
 |
100.0 |
31 |
Fold:Restriction endonuclease-like
Superfamily:Restriction endonuclease-like
Family:YaeQ-like |
| 4 | c3sftA_
|
|
 |
33.8 |
16 |
PDB header:hydrolase
Chain: A: PDB Molecule:chemotaxis response regulator protein-glutamate
PDBTitle: crystal structure of thermotoga maritima cheb methylesterase catalytic2 domain
|
| 5 | d2sh1a_
|
|
 |
32.1 |
25 |
Fold:Defensin-like
Superfamily:Defensin-like
Family:Defensin |
| 6 | d2csua2
|
|
 |
27.8 |
14 |
Fold:Flavodoxin-like
Superfamily:Succinyl-CoA synthetase domains
Family:Succinyl-CoA synthetase domains |
| 7 | d1chda_
|
|
 |
24.0 |
7 |
Fold:Methylesterase CheB, C-terminal domain
Superfamily:Methylesterase CheB, C-terminal domain
Family:Methylesterase CheB, C-terminal domain |
| 8 | c3be3A_
|
|
 |
23.5 |
47 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of a protein belonging to pfam duf16532 from bordetella bronchiseptica
|
| 9 | c3pu5A_
|
|
 |
21.0 |
4 |
PDB header:transport protein
Chain: A: PDB Molecule:extracellular solute-binding protein;
PDBTitle: the crystal structure of a putative extracellular solute-binding2 protein from bordetella parapertussis
|
| 10 | c1a2oB_
|
|
 |
19.2 |
4 |
PDB header:bacterial chemotaxis
Chain: B: PDB Molecule:cheb methylesterase;
PDBTitle: structural basis for methylesterase cheb regulation by a2 phosphorylation-activated domain
|
| 11 | c1uarA_
|
|
 |
16.0 |
7 |
PDB header:transferase
Chain: A: PDB Molecule:rhodanese;
PDBTitle: crystal structure of rhodanese from thermus thermophilus hb8
|
| 12 | d1qvpa_
|
|
 |
13.8 |
16 |
Fold:SH3-like barrel
Superfamily:C-terminal domain of transcriptional repressors
Family:FeoA-like |
| 13 | d1ekqa_
|
|
 |
13.6 |
12 |
Fold:Ribokinase-like
Superfamily:Ribokinase-like
Family:Thiamin biosynthesis kinases |
| 14 | c3hzuA_
|
|
 |
11.6 |
7 |
PDB header:transferase
Chain: A: PDB Molecule:thiosulfate sulfurtransferase ssea;
PDBTitle: crystal structure of probable thiosulfate sulfurtransferase ssea2 (rhodanese) from mycobacterium tuberculosis
|
| 15 | c1e0cA_
|
|
 |
11.3 |
12 |
PDB header:sulfurtransferase
Chain: A: PDB Molecule:sulfurtransferase;
PDBTitle: sulfurtransferase from azotobacter vinelandii
|
| 16 | d1urha2
|
|
 |
11.0 |
5 |
Fold:Rhodanese/Cell cycle control phosphatase
Superfamily:Rhodanese/Cell cycle control phosphatase
Family:Multidomain sulfurtransferase (rhodanese) |
| 17 | c3aaxB_
|
|
 |
10.9 |
14 |
PDB header:transferase
Chain: B: PDB Molecule:putative thiosulfate sulfurtransferase;
PDBTitle: crystal structure of probable thiosulfate sulfurtransferase2 cysa3 (rv3117) from mycobacterium tuberculosis: monoclinic3 form
|
| 18 | c1urhA_
|
|
 |
10.7 |
5 |
PDB header:transferase
Chain: A: PDB Molecule:3-mercaptopyruvate sulfurtransferase;
PDBTitle: the "rhodanese" fold and catalytic mechanism of2 3-mercaptopyruvate sulfotransferases: crystal structure3 of ssea from escherichia coli
|
| 19 | c2p10D_
|
|
 |
10.0 |
23 |
PDB header:hydrolase
Chain: D: PDB Molecule:mll9387 protein;
PDBTitle: crystal structure of a putative phosphonopyruvate hydrolase (mll9387)2 from mesorhizobium loti maff303099 at 2.15 a resolution
|
| 20 | c3olhA_
|
|
 |
9.9 |
3 |
PDB header:transferase
Chain: A: PDB Molecule:3-mercaptopyruvate sulfurtransferase;
PDBTitle: human 3-mercaptopyruvate sulfurtransferase
|
| 21 | d1mjsa_ |
|
not modelled |
9.8 |
37 |
Fold:SMAD/FHA domain
Superfamily:SMAD/FHA domain
Family:SMAD domain |
| 22 | c2kvoA_ |
|
not modelled |
8.7 |
33 |
PDB header:photosynthesis
Chain: A: PDB Molecule:photosystem ii reaction center psb28 protein;
PDBTitle: solution nmr structure of photosystem ii reaction center psb28 protein2 from synechocystis sp.(strain pcc 6803), northeast structural3 genomics consortium target sgr171
|
| 23 | d1khua_ |
|
not modelled |
8.4 |
42 |
Fold:SMAD/FHA domain
Superfamily:SMAD/FHA domain
Family:SMAD domain |
| 24 | d2hy5b1 |
|
not modelled |
8.1 |
21 |
Fold:DsrEFH-like
Superfamily:DsrEFH-like
Family:DsrEF-like |
| 25 | d1wija_ |
|
not modelled |
6.9 |
36 |
Fold:LEM/SAP HeH motif
Superfamily:DNA-binding domain of EIN3-like
Family:DNA-binding domain of EIN3-like |
| 26 | c3pg6D_ |
|
not modelled |
6.7 |
27 |
PDB header:ligase
Chain: D: PDB Molecule:e3 ubiquitin-protein ligase dtx3l;
PDBTitle: the carboxyl terminal domain of human deltex 3-like
|
| 27 | c3eytA_ |
|
not modelled |
6.6 |
13 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein spoa0173;
PDBTitle: crystal structure of thioredoxin-like superfamily protein spoa0173
|
| 28 | d1oi0a_ |
|
not modelled |
6.4 |
14 |
Fold:Cytidine deaminase-like
Superfamily:JAB1/MPN domain
Family:JAB1/MPN domain |
| 29 | d1uara2 |
|
not modelled |
6.3 |
9 |
Fold:Rhodanese/Cell cycle control phosphatase
Superfamily:Rhodanese/Cell cycle control phosphatase
Family:Multidomain sulfurtransferase (rhodanese) |
| 30 | c2oq2B_ |
|
not modelled |
6.2 |
6 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:phosphoadenosine phosphosulfate reductase;
PDBTitle: crystal structure of yeast paps reductase with pap, a product complex
|
| 31 | d2ok5a4 |
|
not modelled |
5.7 |
21 |
Fold:Complement control module/SCR domain
Superfamily:Complement control module/SCR domain
Family:Complement control module/SCR domain |
| 32 | c3ddrC_ |
|
not modelled |
5.6 |
23 |
PDB header:membrane protein/heme binding protein
Chain: C: PDB Molecule:hemophore hasa;
PDBTitle: structure of the serratia marcescens hemophore receptor hasr-ile671gly2 mutant in complex with its hemophore hasa and heme
|
| 33 | c2z5bB_ |
|
not modelled |
5.6 |
19 |
PDB header:chaperone
Chain: B: PDB Molecule:uncharacterized protein ylr021w;
PDBTitle: crystal structure of a novel chaperone complex for yeast2 20s proteasome assembly
|
| 34 | d1hpla2 |
|
not modelled |
5.5 |
5 |
Fold:alpha/beta-Hydrolases
Superfamily:alpha/beta-Hydrolases
Family:Pancreatic lipase, N-terminal domain |
| 35 | c3gztF_ |
|
not modelled |
5.5 |
18 |
PDB header:virus
Chain: F: PDB Molecule:outer capsid glycoprotein vp7;
PDBTitle: vp7 recoated rotavirus dlp
|
| 36 | d2cu2a2 |
|
not modelled |
5.3 |
25 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:mannose-1-phosphate guanylyl transferase |
| 37 | c1boiA_ |
|
not modelled |
5.3 |
5 |
PDB header:transferase
Chain: A: PDB Molecule:rhodanese;
PDBTitle: n-terminally truncated rhodanese
|
| 38 | d1gpla2 |
|
not modelled |
5.1 |
8 |
Fold:alpha/beta-Hydrolases
Superfamily:alpha/beta-Hydrolases
Family:Pancreatic lipase, N-terminal domain |
| 39 | d1lpbb2 |
|
not modelled |
5.0 |
5 |
Fold:alpha/beta-Hydrolases
Superfamily:alpha/beta-Hydrolases
Family:Pancreatic lipase, N-terminal domain |