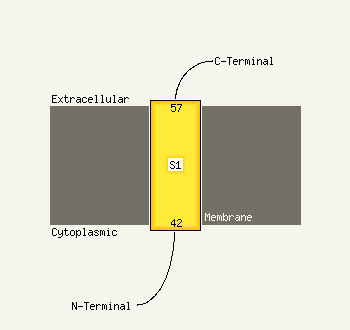
1 c1vzyA_
100.0
24
PDB header: chaperoneChain: A: PDB Molecule: 33 kda chaperonin;PDBTitle: crystal structure of the bacillus subtilis hsp33
2 c1vq0A_
100.0
24
PDB header: chaperoneChain: A: PDB Molecule: 33 kda chaperonin;PDBTitle: crystal structure of 33 kda chaperonin (heat shock protein 33 homolog)2 (hsp33) (tm1394) from thermotoga maritima at 2.20 a resolution
3 c1hw7A_
100.0
100
PDB header: chaperoneChain: A: PDB Molecule: heat shock protein hsp33;PDBTitle: hsp33, heat shock protein with redox-regulated chaperone activity
4 d1hw7a_
100.0
100
Fold: Hsp33 domainSuperfamily: Hsp33 domainFamily: Hsp33 domain5 c1i7fA_
100.0
97
PDB header: chaperoneChain: A: PDB Molecule: heat shock protein 33;PDBTitle: crystal structure of the hsp33 domain with constitutive chaperone2 activity
6 d1vzya1
100.0
24
Fold: Hsp33 domainSuperfamily: Hsp33 domainFamily: Hsp33 domain7 d1vq0a1
100.0
23
Fold: Hsp33 domainSuperfamily: Hsp33 domainFamily: Hsp33 domain8 d1xjha_
99.8
100
Fold: HSP33 redox switch-likeSuperfamily: HSP33 redox switch-likeFamily: HSP33 redox switch-like9 d1vzya2
99.7
30
Fold: HSP33 redox switch-likeSuperfamily: HSP33 redox switch-likeFamily: HSP33 redox switch-like10 d1vq0a2
99.6
28
Fold: HSP33 redox switch-likeSuperfamily: HSP33 redox switch-likeFamily: HSP33 redox switch-like11 c2jz8A_
67.4
27
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein bh09830;PDBTitle: solution nmr structure of bh09830 from bartonella henselae2 modeled with one zn+2 bound. northeast structural genomics3 consortium target bnr55
12 c2h3eB_
59.3
24
PDB header: transferaseChain: B: PDB Molecule: aspartate carbamoyltransferase regulatory chain;PDBTitle: structure of wild-type e. coli aspartate transcarbamoylase in the2 presence of n-phosphonacetyl-l-isoasparagine at 2.3a resolution
13 d2fzcb2
54.2
21
Fold: Rubredoxin-likeSuperfamily: Aspartate carbamoyltransferase, Regulatory-chain, C-terminal domainFamily: Aspartate carbamoyltransferase, Regulatory-chain, C-terminal domain14 d1ywsa1
50.4
24
Fold: Rubredoxin-likeSuperfamily: CSL zinc fingerFamily: CSL zinc finger15 c1pg5B_
48.2
19
PDB header: transferaseChain: B: PDB Molecule: aspartate carbamoyltransferase regulatory chain;PDBTitle: crystal structure of the unligated (t-state) aspartate2 transcarbamoylase from the extremely thermophilic archaeon sulfolobus3 acidocaldarius
16 c2jrrA_
47.8
29
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: solution nmr structure of q5lls5 from silicibacter2 pomeroyi. northeast structural genomics consortium target3 sir90
17 d1pg5b2
44.6
19
Fold: Rubredoxin-likeSuperfamily: Aspartate carbamoyltransferase, Regulatory-chain, C-terminal domainFamily: Aspartate carbamoyltransferase, Regulatory-chain, C-terminal domain18 c2eluA_
40.8
32
PDB header: transcriptionChain: A: PDB Molecule: zinc finger protein 406;PDBTitle: solution structure of the 5th c2h2 zinc finger of human2 zinc finger protein 406
19 c2zaeB_
38.8
16
PDB header: hydrolaseChain: B: PDB Molecule: ribonuclease p protein component 4;PDBTitle: crystal structure of protein ph1601p in complex with protein ph1771p2 of archaeal ribonuclease p from pyrococcus horikoshii ot3
20 c1y6uA_
37.3
11
PDB header: dna binding proteinChain: A: PDB Molecule: excisionase from transposon tn916;PDBTitle: the structure of the excisionase (xis) protein from2 conjugative transposon tn916 provides insights into the3 regulation of heterobivalent tyrosine recombinases
21 c2jvmA_
not modelled
36.3
31
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: solution nmr structure of rhodobacter sphaeroides protein2 rhos4_26430. northeast structural genomics consortium3 target rhr95
22 d1zu1a1
not modelled
34.4
21
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: HkH motif-containing C2H2 finger23 d1nzja_
not modelled
31.9
32
Fold: Adenine nucleotide alpha hydrolase-likeSuperfamily: Nucleotidylyl transferaseFamily: Class I aminoacyl-tRNA synthetases (RS), catalytic domain24 d1gtra2
not modelled
30.7
5
Fold: Adenine nucleotide alpha hydrolase-likeSuperfamily: Nucleotidylyl transferaseFamily: Class I aminoacyl-tRNA synthetases (RS), catalytic domain25 d2atcb2
not modelled
27.9
25
Fold: Rubredoxin-likeSuperfamily: Aspartate carbamoyltransferase, Regulatory-chain, C-terminal domainFamily: Aspartate carbamoyltransferase, Regulatory-chain, C-terminal domain26 c2eodA_
not modelled
27.9
24
PDB header: signaling proteinChain: A: PDB Molecule: tnf receptor-associated factor 4;PDBTitle: solution structure of traf-type zinc finger domains (190-2 248) from human tnf receptor-associated factor 4
27 c2be7E_
not modelled
27.5
25
PDB header: transferaseChain: E: PDB Molecule: aspartate carbamoyltransferase regulatory chain;PDBTitle: crystal structure of the unliganded (t-state) aspartate2 transcarbamoylase of the psychrophilic bacterium moritella profunda
28 d1wiia_
not modelled
27.0
10
Fold: Rubredoxin-likeSuperfamily: Zinc beta-ribbonFamily: Putative zinc binding domain29 c2l8eA_
not modelled
26.6
44
PDB header: dna binding proteinChain: A: PDB Molecule: polyhomeotic-like protein 1;PDBTitle: solution nmr structure of fcs domain of human polyhomeotic homolog 12 (hph1)
30 c3mv2A_
not modelled
26.0
13
PDB header: protein transportChain: A: PDB Molecule: coatomer subunit alpha;PDBTitle: crystal structure of a-cop in complex with e-cop
31 c3q9qB_
not modelled
26.0
16
PDB header: chaperoneChain: B: PDB Molecule: heat shock protein beta-1;PDBTitle: hspb1 fragment second crystal form
32 c3t7vA_
not modelled
25.2
27
PDB header: transferaseChain: A: PDB Molecule: methylornithine synthase pylb;PDBTitle: crystal structure of methylornithine synthase (pylb)
33 c2xvcA_
not modelled
23.8
8
PDB header: cell cycleChain: A: PDB Molecule: escrt-iii;PDBTitle: molecular and structural basis of escrt-iii recruitment to2 membranes during archaeal cell division
34 d1j09a2
not modelled
23.2
10
Fold: Adenine nucleotide alpha hydrolase-likeSuperfamily: Nucleotidylyl transferaseFamily: Class I aminoacyl-tRNA synthetases (RS), catalytic domain35 c2elpA_
not modelled
22.1
29
PDB header: transcriptionChain: A: PDB Molecule: zinc finger protein 406;PDBTitle: solution structure of the 13th c2h2 zinc finger of human2 zinc finger protein 406
36 d1wgea1
not modelled
21.2
16
Fold: Rubredoxin-likeSuperfamily: CSL zinc fingerFamily: CSL zinc finger37 c3mkrB_
not modelled
20.8
27
PDB header: transport proteinChain: B: PDB Molecule: coatomer subunit alpha;PDBTitle: crystal structure of yeast alpha/epsilon-cop subcomplex of the copi2 vesicular coat
38 c2wulB_
not modelled
20.8
17
PDB header: oxidoreductaseChain: B: PDB Molecule: glutaredoxin related protein 5;PDBTitle: crystal structure of the human glutaredoxin 5 with bound2 glutathione in an fes cluster
39 c2ywwA_
not modelled
20.8
19
PDB header: metal binding proteinChain: A: PDB Molecule: aspartate carbamoyltransferase regulatory chain;PDBTitle: crystal structure of aspartate carbamoyltransferase2 regulatory chain from methanocaldococcus jannaschii
40 c2d0oA_
not modelled
20.8
15
PDB header: chaperoneChain: A: PDB Molecule: diol dehydratase-reactivating factor largePDBTitle: strcuture of diol dehydratase-reactivating factor complexed2 with adp and mg2+
41 c1wysA_
not modelled
20.8
30
PDB header: metal binding proteinChain: A: PDB Molecule: riken cdna 2310008m20 protein;PDBTitle: solution structure of the first zf-an1 domain of mouse2 riken cdna 2310008m20 protein
42 d2yrka1
not modelled
20.7
36
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: HkH motif-containing C2H2 finger43 c2k5rA_
not modelled
20.6
0
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein xf2673;PDBTitle: solution nmr structure of xf2673 from xylella fastidiosa.2 northeast structural genomics consortium target xfr39
44 c1pqvS_
not modelled
20.1
30
PDB header: transferase/transcriptionChain: S: PDB Molecule: transcription elongation factor s-ii;PDBTitle: rna polymerase ii-tfiis complex
45 c1y1yS_
not modelled
20.1
30
PDB header: transferase/transcription/dna-rna hybridChain: S: PDB Molecule: transcription elongation factor s-ii;PDBTitle: rna polymerase ii-tfiis-dna/rna complex
46 d1gx5a_
not modelled
19.4
17
Fold: DNA/RNA polymerasesSuperfamily: DNA/RNA polymerasesFamily: RNA-dependent RNA-polymerase47 c2xigA_
not modelled
18.9
21
PDB header: transcriptionChain: A: PDB Molecule: ferric uptake regulation protein;PDBTitle: the structure of the helicobacter pylori ferric uptake2 regulator fur reveals three functional metal binding sites
48 c2w0tA_
not modelled
18.9
29
PDB header: transcriptionChain: A: PDB Molecule: lethal(3)malignant brain tumor-like 2 protein;PDBTitle: solution structure of the fcs zinc finger domain of human2 lmbl2
49 d1vd4a_
not modelled
18.1
11
Fold: Rubredoxin-likeSuperfamily: Zinc beta-ribbonFamily: Transcriptional factor domain50 c2l6lA_
not modelled
17.7
24
PDB header: chaperoneChain: A: PDB Molecule: dnaj homolog subfamily c member 24;PDBTitle: solution structure of human j-protein co-chaperone, dph4
51 c3iufA_
not modelled
17.5
41
PDB header: protein bindingChain: A: PDB Molecule: zinc finger protein ubi-d4;PDBTitle: crystal structure of the c2h2-type zinc finger domain of2 human ubi-d4
52 d1hxra_
not modelled
17.3
33
Fold: Mss4-likeSuperfamily: Mss4-likeFamily: RabGEF Mss453 d1wfea_
not modelled
17.3
30
Fold: AN1-like Zinc fingerSuperfamily: AN1-like Zinc fingerFamily: AN1-like Zinc finger54 d1nb4a_
not modelled
16.9
17
Fold: DNA/RNA polymerasesSuperfamily: DNA/RNA polymerasesFamily: RNA-dependent RNA-polymerase55 c3cixA_
not modelled
16.5
45
PDB header: adomet binding proteinChain: A: PDB Molecule: fefe-hydrogenase maturase;PDBTitle: x-ray structure of the [fefe]-hydrogenase maturase hyde from2 thermotoga maritima in complex with thiocyanate
56 d2jnya1
not modelled
16.3
8
Fold: Trm112p-likeSuperfamily: Trm112p-likeFamily: Trm112p-like57 c1zu1A_
not modelled
16.3
27
PDB header: rna binding proteinChain: A: PDB Molecule: rna binding protein zfa;PDBTitle: solution structure of the n-terminal zinc fingers of the2 xenopus laevis double stranded rna binding protein zfa
58 c2j6aA_
not modelled
16.2
33
PDB header: transferaseChain: A: PDB Molecule: protein trm112;PDBTitle: crystal structure of s. cerevisiae ynr046w, a zinc finger2 protein from the erf1 methyltransferase complex.
59 d2dkta1
not modelled
15.7
14
Fold: CHY zinc finger-likeSuperfamily: CHY zinc finger-likeFamily: CHY zinc finger60 c1xf7A_
not modelled
15.4
22
PDB header: transcriptionChain: A: PDB Molecule: wilms' tumor protein;PDBTitle: high resolution nmr structure of the wilms' tumor2 suppressor protein (wt1) finger 3
61 d1xf7a_
not modelled
15.4
22
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Classic zinc finger, C2H262 d2fu5a1
not modelled
15.3
33
Fold: Mss4-likeSuperfamily: Mss4-likeFamily: RabGEF Mss463 d2pk7a1
not modelled
15.3
15
Fold: Trm112p-likeSuperfamily: Trm112p-likeFamily: Trm112p-like64 c1rikA_
not modelled
15.0
22
PDB header: de novo proteinChain: A: PDB Molecule: e6apc1 peptide;PDBTitle: e6-binding zinc finger (e6apc1)
65 c1arfA_
not modelled
14.5
22
PDB header: transcription regulationChain: A: PDB Molecule: yeast transcription factor adr1;PDBTitle: structures of dna-binding mutant zinc finger domains:2 implications for dna binding
66 d1llmc2
not modelled
14.5
44
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Classic zinc finger, C2H267 d1tkea2
not modelled
14.4
7
Fold: RRF/tRNA synthetase additional domain-likeSuperfamily: ThrRS/AlaRS common domainFamily: Threonyl-tRNA synthetase (ThrRS), second 'additional' domain68 c3py7A_
not modelled
14.4
23
PDB header: viral proteinChain: A: PDB Molecule: maltose-binding periplasmic protein,paxillin ld1,protein e6PDBTitle: crystal structure of full-length bovine papillomavirus oncoprotein e62 in complex with ld1 motif of paxillin at 2.3a resolution
69 d1wima_
not modelled
14.3
14
Fold: RING/U-boxSuperfamily: RING/U-boxFamily: RING finger domain, C3HC470 d1wpba_
not modelled
14.2
33
Fold: YfbU-likeSuperfamily: YfbU-likeFamily: YfbU-like71 d2dmda2
not modelled
13.9
50
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Classic zinc finger, C2H272 c2js4A_
not modelled
13.9
8
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: upf0434 protein bb2007;PDBTitle: solution nmr structure of bordetella bronchiseptica protein2 bb2007. northeast structural genomics consortium target3 bor54
73 c2hz7A_
not modelled
13.5
14
PDB header: ligaseChain: A: PDB Molecule: glutaminyl-trna synthetase;PDBTitle: crystal structure of the glutaminyl-trna synthetase from2 deinococcus radiodurans
74 c2jr7A_
not modelled
13.4
16
PDB header: metal binding proteinChain: A: PDB Molecule: dph3 homolog;PDBTitle: solution structure of human desr1
75 d2hf1a1
not modelled
13.2
8
Fold: Trm112p-likeSuperfamily: Trm112p-likeFamily: Trm112p-like76 d1k81a_
not modelled
12.8
23
Fold: Zinc-binding domain of translation initiation factor 2 betaSuperfamily: Zinc-binding domain of translation initiation factor 2 betaFamily: Zinc-binding domain of translation initiation factor 2 beta77 d1a1ia3
not modelled
12.8
44
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Classic zinc finger, C2H278 d2yt9a2
not modelled
12.8
27
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Classic zinc finger, C2H279 c3ol0C_
not modelled
12.8
23
PDB header: de novo proteinChain: C: PDB Molecule: de novo designed monomer trefoil-fold sub-domain whichPDBTitle: crystal structure of monofoil-4p homo-trimer: de novo designed monomer2 trefoil-fold sub-domain which forms homo-trimer assembly
80 d1njqa_
not modelled
12.7
22
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Plant C2H2 finger (QALGGH zinc finger)81 c3iraA_
not modelled
12.7
21
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: conserved protein;PDBTitle: the crystal structure of one domain of the conserved protein from2 methanosarcina mazei go1
82 d1vbka2
not modelled
12.6
8
Fold: THUMP domainSuperfamily: THUMP domain-likeFamily: THUMP domain83 c2d9kA_
not modelled
12.6
13
PDB header: immune systemChain: A: PDB Molecule: fln29 gene product;PDBTitle: solution structure of the zf-traf domain of fln29 gene2 product
84 c3h5jA_
not modelled
12.5
32
PDB header: lyaseChain: A: PDB Molecule: 3-isopropylmalate dehydratase small subunit;PDBTitle: leud_1-168 small subunit of isopropylmalate isomerase (rv2987c) from2 mycobacterium tuberculosis
85 d2ds5a1
not modelled
12.5
38
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: ClpX chaperone zinc binding domain86 c2pjhA_
not modelled
12.3
12
PDB header: transport proteinChain: A: PDB Molecule: nuclear protein localization protein 4 homolog;PDBTitle: strctural model of the p97 n domain- npl4 ubd complex
87 d1k3sa_
not modelled
12.2
15
Fold: Secretion chaperone-likeSuperfamily: Type III secretory system chaperone-likeFamily: Type III secretory system chaperone88 c1ovxB_
not modelled
12.1
38
PDB header: metal binding proteinChain: B: PDB Molecule: atp-dependent clp protease atp-binding subunit clpx;PDBTitle: nmr structure of the e. coli clpx chaperone zinc binding domain dimer
89 c3e2iA_
not modelled
12.1
14
PDB header: transferaseChain: A: PDB Molecule: thymidine kinase;PDBTitle: crystal structure of thymidine kinase from s. aureus
90 c2hlgA_
not modelled
12.0
29
PDB header: plant proteinChain: A: PDB Molecule: fruit-specific protein;PDBTitle: nmr solution structure of a new tomato peptide
91 c4a17Y_
not modelled
11.9
30
PDB header: ribosomeChain: Y: PDB Molecule: rpl37a;PDBTitle: t.thermophila 60s ribosomal subunit in complex with2 initiation factor 6. this file contains 5s rrna,3 5.8s rrna and proteins of molecule 2.
92 d2ct1a2
not modelled
11.9
22
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Classic zinc finger, C2H293 c1x4vA_
not modelled
11.9
20
PDB header: metal binding proteinChain: A: PDB Molecule: hypothetical protein loc130617;PDBTitle: solution structure of the zf-an1 domain from human2 hypothetical protein loc130617
94 d1kjwa2
not modelled
11.8
13
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nucleotide and nucleoside kinases95 d1e4ft2
not modelled
11.6
8
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: Actin/HSP7096 c3aiiA_
not modelled
11.5
27
PDB header: ligaseChain: A: PDB Molecule: glutamyl-trna synthetase;PDBTitle: archaeal non-discriminating glutamyl-trna synthetase from2 methanothermobacter thermautotrophicus
97 d1nj1a2
not modelled
11.5
33
Fold: IF3-likeSuperfamily: C-terminal domain of ProRSFamily: C-terminal domain of ProRS98 c1tr8A_
not modelled
11.4
23
PDB header: chaperoneChain: A: PDB Molecule: conserved protein (mth177);PDBTitle: crystal structure of archaeal nascent polypeptide-associated complex2 (aenac)
99 c2hcuA_
not modelled
11.3
37
PDB header: lyaseChain: A: PDB Molecule: 3-isopropylmalate dehydratase small subunit;PDBTitle: crystal structure of smu.1381 (or leud) from streptococcus2 mutans