| 1 |
|
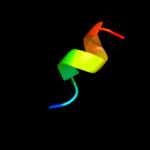
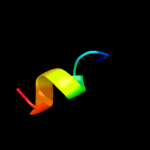
PDB 2e74 chain B domain 1
Region: 2 - 11
Aligned: 10
Modelled: 10
Confidence: 22.4%
Identity: 50%
Fold: a domain/subunit of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)
Superfamily: a domain/subunit of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)
Family: a domain/subunit of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)
Phyre2
| 2 |
|
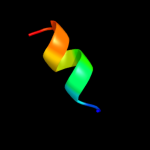
PDB 1q90 chain D
Region: 2 - 11
Aligned: 10
Modelled: 10
Confidence: 21.8%
Identity: 40%
Fold: a domain/subunit of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)
Superfamily: a domain/subunit of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)
Family: a domain/subunit of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)
Phyre2
| 3 |
|
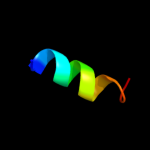
PDB 1bcc chain C domain 2
Region: 2 - 11
Aligned: 10
Modelled: 10
Confidence: 18.6%
Identity: 50%
Fold: a domain/subunit of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)
Superfamily: a domain/subunit of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)
Family: a domain/subunit of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)
Phyre2
| 4 |
|
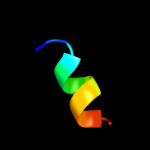
PDB 1ppj chain C domain 1
Region: 2 - 11
Aligned: 10
Modelled: 10
Confidence: 16.5%
Identity: 50%
Fold: a domain/subunit of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)
Superfamily: a domain/subunit of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)
Family: a domain/subunit of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)
Phyre2
| 5 |
|
PDB 3cx5 chain C domain 1
Region: 2 - 11
Aligned: 10
Modelled: 10
Confidence: 16.4%
Identity: 70%
Fold: a domain/subunit of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)
Superfamily: a domain/subunit of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)
Family: a domain/subunit of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)
Phyre2
| 6 |
|
PDB 2ys9 chain A
Region: 20 - 28
Aligned: 9
Modelled: 9
Confidence: 16.2%
Identity: 67%
PDB header:transcription
Chain: A: PDB Molecule:homeobox and leucine zipper protein homez;
PDBTitle: structure of the third homeodomain from the human homeobox2 and leucine zipper protein, homez
Phyre2
| 7 |
|
PDB 1vf5 chain B
Region: 2 - 11
Aligned: 10
Modelled: 10
Confidence: 16.0%
Identity: 50%
Fold: a domain/subunit of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)
Superfamily: a domain/subunit of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)
Family: a domain/subunit of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)
Phyre2
| 8 |
|
PDB 1z65 chain A
Region: 6 - 19
Aligned: 14
Modelled: 14
Confidence: 14.7%
Identity: 50%
PDB header:unknown function
Chain: A: PDB Molecule:prion-like protein doppel;
PDBTitle: mouse doppel 1-30 peptide
Phyre2
| 9 |
|
PDB 1tj1 chain A domain 1
Region: 9 - 14
Aligned: 6
Modelled: 6
Confidence: 11.5%
Identity: 67%
Fold: N-terminal domain of bifunctional PutA protein
Superfamily: N-terminal domain of bifunctional PutA protein
Family: N-terminal domain of bifunctional PutA protein
Phyre2
| 10 |
|
PDB 3cx5 chain N
Region: 2 - 11
Aligned: 10
Modelled: 10
Confidence: 6.9%
Identity: 70%
PDB header:oxidoreductase
Chain: N: PDB Molecule:cytochrome b;
PDBTitle: structure of complex iii with bound cytochrome c in reduced2 state and definition of a minimal core interface for3 electron transfer.
Phyre2
| 11 |
|
PDB 2qjk chain M
Region: 2 - 11
Aligned: 10
Modelled: 10
Confidence: 6.8%
Identity: 60%
PDB header:electron transport
Chain: M: PDB Molecule:cytochrome b;
PDBTitle: crystal structure analysis of mutant rhodobacter2 sphaeroides bc1 with stigmatellin and antimycin
Phyre2
| 12 |
|
PDB 3cwb chain C
Region: 2 - 11
Aligned: 10
Modelled: 10
Confidence: 6.4%
Identity: 50%
PDB header:oxidoreductase
Chain: C: PDB Molecule:cytochrome b;
PDBTitle: chicken cytochrome bc1 complex inhibited by an iodinated analogue of2 the polyketide crocacin-d
Phyre2
| 13 |
|
PDB 2kne chain B
Region: 4 - 10
Aligned: 7
Modelled: 7
Confidence: 6.0%
Identity: 57%
PDB header:metal transport
Chain: B: PDB Molecule:atpase, ca++ transporting, plasma membrane 4;
PDBTitle: calmodulin wraps around its binding domain in the plasma2 membrane ca2+ pump anchored by a novel 18-1 motif
Phyre2
| 14 |
|
PDB 2wfv chain A
Region: 14 - 19
Aligned: 6
Modelled: 6
Confidence: 5.8%
Identity: 83%
PDB header:signaling protein
Chain: A: PDB Molecule:probable insulin-like peptide 5 a chain;
PDBTitle: crystal structure of dilp5 variant c4
Phyre2
| 15 |
|
PDB 2p7t chain C domain 1
Region: 6 - 9
Aligned: 4
Modelled: 4
Confidence: 5.8%
Identity: 100%
Fold: Voltage-gated potassium channels
Superfamily: Voltage-gated potassium channels
Family: Voltage-gated potassium channels
Phyre2
| 16 |
|
PDB 2wfu chain A
Region: 14 - 19
Aligned: 6
Modelled: 6
Confidence: 5.8%
Identity: 83%
PDB header:signaling protein
Chain: A: PDB Molecule:probable insulin-like peptide 5 a chain;
PDBTitle: crystal structure of dilp5 variant db
Phyre2