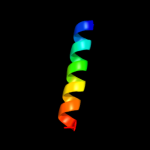| Secondary structure and disorder prediction | |
 |
| | |
1 | . | . | . | . | . | . | . | . | 10 | . | . | . | . | . | . | . | . | . | 20 | . | . | . | . | . | . | . | . | . | 30 | . | . | . | . | . | . | . | . | . | 40 | . | . | . | . | . | . | . | . | . | 50 | . | . | . | . | . | . | . | . | . | 60 |
| Sequence | |
M | S | K | A | G | K | I | T | A | A | I | S | G | A | F | L | L | L | I | V | V | A | I | I | L | I | A | T | F | D | W | N | R | L | K | P | T | I | N | Q | K | V | S | A | E | L | N | R | P | F | A | I | R | G | D | L | G | V | V | W |
| Secondary structure | |
|
|  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |
|
|
|  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |
|
|  |  |  |  |  |  |  |  |  |  |  |  |
| SS confidence | |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| Disorder | |
? | ? | ? | ? |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| Disorder confidence | |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| |
| | |
. | . | . | . | . | . | . | . | . | 70 | . | . | . | . | . | . | . | . | . | 80 | . | . | . | . | . | . | . | . | . | 90 | . | . | . | . | . | . | . | . | . | 100 | . | . | . | . | . | . | . | . | . | 110 | . | . | . | . | . | . | . | . | . | 120 |
| Sequence | |
E | R | Q | K | Q | E | T | G | W | R | S | W | V | P | W | P | H | V | H | A | E | D | I | I | L | G | N | P | P | D | I | P | E | V | T | M | V | H | L | P | R | V | E | A | T | L | A | P | L | A | L | L | T | K | T | V | W | L | P | W |
| Secondary structure | |
 |
|
|
|
|
|
|
|  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |
|
|
|
|
|
|
|
|
|  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |
|
|
|  |  |  |  |  |
| SS confidence | |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| Disorder | |
| ? | ? | ? | ? | ? | ? | ? | ? |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| ? | ? | ? | ? | ? | ? |
| ? |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| ? |
| ? |
|
|
|
|
|
|
|
| Disorder confidence | |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| |
| | |
. | . | . | . | . | . | . | . | . | 130 | . | . | . | . | . | . | . | . | . | 140 | . | . | . | . | . | . | . | . | . | 150 | . | . | . | . | . | . | . | . | . | 160 | . | . | . | . | . | . | . | . | . | 170 | . | . | . | . | . | . | . | . | . | 180 |
| Sequence | |
I | K | L | E | K | P | D | A | R | L | I | R | L | S | E | K | N | N | N | W | T | F | N | L | A | N | D | D | N | K | D | A | N | A | K | P | S | A | W | S | F | R | L | D | N | I | L | F | D | Q | G | R | I | A | I | D | D | K | V | S |
| Secondary structure | |
 |  |  |  |
|
|  |  |  |  |  |  |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|  |  |  |  |
|
|  |  |  |
|
|  |  |  |  |  |
|
|
|
|
| SS confidence | |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| Disorder | |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| ? |
|
|
|
| ? | ? | ? | ? | ? | ? | ? | ? | ? | ? | ? | ? | ? | ? | ? | ? | ? |
|
|
|
|
| ? |
| ? |
|
|
|
|
|
|
|
|
|
|
| ? |
| ? |
| Disorder confidence | |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| |
| | |
. | . | . | . | . | . | . | . | . | 190 | . | . | . | . | . | . | . | . | . | 200 | . | . | . | . | . | . | . | . | . | 210 | . | . | . | . | . | . | . | . | . | 220 | . | . | . | . | . | . | . | . | . | 230 | . | . | . | . | . | . | . | . | . | 240 |
| Sequence | |
K | A | D | L | E | I | F | V | D | P | L | G | K | P | L | P | F | S | E | V | T | G | S | K | G | K | A | D | K | E | K | V | G | D | Y | V | F | G | L | K | A | Q | G | R | Y | N | G | E | P | L | T | G | T | G | K | I | G | G | M | L |
| Secondary structure | |
|
|
|
|  |  |  |  |  |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|  |  |  |  |  |  |  |  |  |  |
|
|
|  |  |  |  |  |  |
|
|
|
|
|
|
| SS confidence | |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| Disorder | |
? |
|
|
|
|
|
|
|
|
| ? | ? | ? | ? | ? |
| ? | ? | ? | ? | ? | ? | ? | ? | ? | ? | ? | ? | ? | ? | ? | ? | ? | ? | ? |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| ? |
|
|
|
| ? |
|
|
|
| Disorder confidence | |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| |
| | |
. | . | . | . | . | . | . | . | . | 250 | . | . | . | . | . | . | . | . | . | 260 | . | . | . | . | . | . | . | . | . | 270 | . | . | . | . | . | . | . | . | . | 280 | . | . | . | . | . | . | . | . | . | 290 | . | . | . | . | . | . | . | . | . | 300 |
| Sequence | |
A | L | R | G | E | G | T | P | F | P | V | Q | A | D | F | R | S | G | N | T | R | V | A | F | D | G | V | V | N | D | P | M | K | M | G | G | V | D | L | R | L | K | F | S | G | D | S | L | G | D | L | Y | E | L | T | G | V | L | L | P |
| Secondary structure | |
|
|
|
|
|
|
|  |  |  |  |  |  |  |  |  |
|
|
|  |  |  |  |  |  |  |  |  |
|
|
|
|
|
|
|  |  |  |  |  |  |  |  |  |  |
|
|  |  |  |  |  |  |  |
|
|
|
|
|
|
| SS confidence | |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| Disorder | |
? | ? | ? | ? | ? | ? | ? | ? |
|
|
|
|
|
|
| ? |
|
| ? |
|
|
|
|
|
|
|
|
|
|
| ? | ? |
| ? |
| ? |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| ? |
|
| ? |
| ? |
| Disorder confidence | |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| |
| | |
. | . | . | . | . | . | . | . | . | 310 | . | . | . | . | . | . | . | . | . | 320 | . | . | . | . | . | . | . | . | . | 330 | . | . | . | . | . | . | . | . | . | 340 | . | . | . | . | . | . | . | . | . | 350 | . | . | . | . | . | . | . | . | . | 360 |
| Sequence | |
D | T | P | P | F | E | T | D | G | R | L | V | A | K | I | D | T | E | K | S | S | V | F | D | Y | R | G | F | N | G | R | I | G | D | S | D | I | H | G | S | L | V | Y | T | T | G | K | P | R | P | K | L | E | G | D | V | E | S | R | Q |
| Secondary structure | |
|
|
|
|
|  |  |  |  |  |  |  |  |  |
|
|
|
|
|
|
|  |  |  |  |  |
|
|
|
|  |  |
|
|  |  |  |  |  |  |  |  |  |  |
|
|
|
|
|  |  |  |  |  |  |  |  |
|
|
|
| SS confidence | |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| Disorder | |
? | ? | ? | ? | ? | ? | ? |
|
|
|
|
| ? | ? | ? |
| ? | ? | ? | ? | ? |
|
|
|
|
|
|
| ? |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| ? |
|
|
|
|
|
|
|
|
|
|
|
| Disorder confidence | |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| |
| | |
. | . | . | . | . | . | . | . | . | 370 | . | . | . | . | . | . | . | . | . | 380 | . | . | . | . | . | . | . | . | . | 390 | . | . | . | . | . | . | . | . | . | 400 | . | . | . | . | . | . | . | . | . | 410 | . | . | . | . | . | . | . | . | . | 420 |
| Sequence | |
L | R | L | A | D | L | G | P | L | I | G | V | D | S | G | K | G | A | E | K | S | K | R | S | E | Q | K | K | G | E | K | S | V | Q | P | A | G | K | V | L | P | Y | D | R | F | E | T | D | K | W | D | V | M | D | A | D | V | R | F | K |
| Secondary structure | |
 |  |  |  |  |  |  |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|  |  |
|
|
|  |  |  |  |  |
| SS confidence | |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| Disorder | |
|
|
|
|
|
| ? | ? | ? | ? | ? | ? | ? | ? | ? | ? | ? | ? | ? | ? | ? | ? | ? | ? | ? | ? | ? | ? | ? | ? | ? | ? | ? | ? | ? | ? | ? | ? | ? | ? | ? | ? |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| Disorder confidence | |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| |
| | |
. | . | . | . | . | . | . | . | . | 430 | . | . | . | . | . | . | . | . | . | 440 | . | . | . | . | . | . | . | . | . | 450 | . | . | . | . | . | . | . | . | . | 460 | . | . | . | . | . | . | . | . | . | 470 | . | . | . | . | . | . | . | . | . | 480 |
| Sequence | |
G | R | R | I | E | H | G | S | S | L | P | I | S | D | L | S | T | H | I | I | L | K | N | A | D | L | R | L | Q | P | L | K | F | G | M | A | G | G | S | I | A | A | N | I | H | L | E | G | D | K | K | P | M | Q | G | R | A | D | I | Q |
| Secondary structure | |
 |  |  |  |  |  |
|
|
|
|  |  |  |  |  |  |  |  |  |  |  |  |
|
|  |  |  |  |  |  |  |  |  |  |  |  |
|
|  |  |  |  |  |  |  |  |  |  |
|
|
|
|  |  |  |  |  |  |  |  |
| SS confidence | |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| Disorder | |
|
|
| ? | ? | ? | ? | ? |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| ? | ? |
| ? |
|
|
|
|
|
|
|
|
|
|
|
| ? | ? | ? |
| ? | ? |
|
|
|
|
|
|
|
| Disorder confidence | |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| |
| | |
. | . | . | . | . | . | . | . | . | 490 | . | . | . | . | . | . | . | . | . | 500 | . | . | . | . | . | . | . | . | . | 510 | . | . | . | . | . | . | . | . | . | 520 | . | . | . | . | . | . | . | . | . | 530 | . | . | . | . | . | . | . | . | . | 540 |
| Sequence | |
A | R | R | L | K | L | K | E | L | M | P | D | V | E | L | M | Q | K | T | L | G | E | M | N | G | D | A | E | L | R | G | S | G | N | S | V | A | A | L | L | G | N | S | N | G | N | L | K | L | L | M | N | D | G | L | V | S | R | N | L |
| Secondary structure | |
 |
|
|
|
|  |  |  |  |  |  |  |
|
|
|
|
|  |  |  |  |  |  |  |  |  |  |  |  |  |  |
|
|
|
|  |  |  |  |  |  |  |
|
|
|  |  |  |  |  |  |  |
|
|  |  |  |
|
|  |
| SS confidence | |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| Disorder | |
|
|
|
|
|
|
|
|
|
|
|
| ? | ? | ? | ? | ? | ? | ? | ? |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| ? | ? |
|
| Disorder confidence | |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| |
| | |
. | . | . | . | . | . | . | . | . | 550 | . | . | . | . | . | . | . | . | . | 560 | . | . | . | . | . | . | . | . | . | 570 | . | . | . | . | . | . | . | . | . | 580 | . | . | . | . | . | . | . | . | . | 590 | . | . | . | . | . | . | . | . | . | 600 |
| Sequence | |
M | E | I | V | G | L | N | V | G | N | Y | I | V | G | A | I | F | G | D | D | E | V | R | V | N | C | A | A | A | N | L | N | I | A | N | G | V | A | R | P | Q | I | F | A | F | D | T | E | N | A | L | I | N | V | T | G | T | A | S | F |
| Secondary structure | |
 |  |  |  |  |  |  |  |  |  |  |  |  |  |  |
|
|
|
|
|
|  |  |  |  |  |  |  |  |  |  |  |  |  |
|
|  |  |  |  |
|  |  |  |  |  |  |
|
|  |  |  |  |  |  |  |  |  |  |  |
| SS confidence | |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| Disorder | |
|
|
|
| ? | ? |
|
|
|
|
|
| ? | ? | ? | ? | ? | ? | ? | ? | ? | ? |
| ? | ? |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| ? | ? | ? |
|
|
| ? | ? | ? | ? |
|
|
|
|
|
|
|
|
|
|
| Disorder confidence | |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| |
| | |
. | . | . | . | . | . | . | . | . | 610 | . | . | . | . | . | . | . | . | . | 620 | . | . | . | . | . | . | . | . | . | 630 | . | . | . | . | . | . | . | . | . | 640 | . | . | . | . | . | . | . | . | . | 650 | . | . | . | . | . | . | . | . | . | 660 |
| Sequence | |
A | S | E | Q | L | D | L | T | I | D | P | E | S | K | G | I | R | I | I | T | L | R | S | P | L | Y | V | R | G | T | F | K | N | P | Q | A | G | V | K | A | G | P | L | I | A | R | G | A | V | A | A | A | L | A | T | L | V | T | P | A |
| Secondary structure | |
|
|
|  |  |  |  |  |  |  |  |
|
|
|
|
|
|
|
|
|  |  |  |  |  |  |  |  |  |  |
|
|
|
|  |  |  |  |
|  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |
|
|  |  |
| SS confidence | |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| Disorder | |
|
|
|
|
|
|
|
|
|
|
| ? | ? | ? | ? | ? | ? | ? | ? | ? | ? | ? | ? |
|
|
|
|
|
|
|
|
|
| ? |
|
|
|
| ? |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| ? |
|
|
| Disorder confidence | |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| |
| | |
. | . | . | . | . | . | . | . | . | 670 | . | . | . | . | . | . | . | . | . | 680 | . | . | . | . | . | . |
| Sequence | |
A | A | L | L | A | L | I | S | P | S | E | G | E | A | N | Q | C | R | T | I | L | S | Q | M | K | K |
| Secondary structure | |
 |  |  |  |  |  |
|
|
|
|
|
|
|
|
|
|  |  |  |  |  |  |  |  |
|
|
| SS confidence | |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| Disorder | |
|
|
|
|
| ? | ? | ? | ? | ? | ? | ? | ? | ? | ? | ? |
|
|
|
|
|
|
|
| ? | ? |
| Disorder confidence | |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| |
| Confidence Key |
| High(9) | |
|
|
|
|
|
|
|
|
|
Low (0) |
| ? | Disordered |
  | Alpha helix |
  | Beta strand |
Hover over an aligned region to see model and summary info
Please note, only up to the top 20 hits are modelled to reduce computer load
|
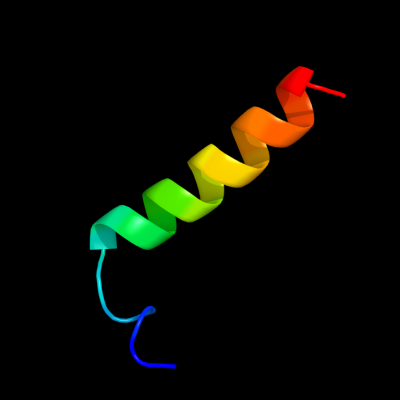
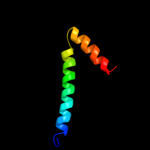
| 1 |
|
PDB 1w8x chain P
Region: 1 - 24
Aligned: 23
Modelled: 24
Confidence: 67.5%
Identity: 22%
PDB header:virus
Chain: P: PDB Molecule:protein p16;
PDBTitle: structural analysis of prd1
Phyre2
| 2 |
|
PDB 1v54 chain G
Region: 1 - 21
Aligned: 21
Modelled: 21
Confidence: 28.4%
Identity: 10%
Fold: Single transmembrane helix
Superfamily: Mitochondrial cytochrome c oxidase subunit VIa
Family: Mitochondrial cytochrome c oxidase subunit VIa
Phyre2
| 3 |
|
PDB 1x4q chain A
Region: 30 - 47
Aligned: 18
Modelled: 18
Confidence: 13.5%
Identity: 33%
PDB header:rna binding protein
Chain: A: PDB Molecule:u4/u6 small nuclear ribonucleoprotein prp3;
PDBTitle: solution structure of pwi domain in u4/u6 small nuclear2 ribonucleoprotein prp3(hprp3)
Phyre2
| 4 |
|
PDB 1wx4 chain B
Region: 120 - 138
Aligned: 19
Modelled: 19
Confidence: 8.3%
Identity: 5%
PDB header:oxidoreductase/metal transport
Chain: B: PDB Molecule:melc;
PDBTitle: crystal structure of the oxy-form of the copper-bound streptomyces2 castaneoglobisporus tyrosinase complexed with a caddie protein3 prepared by the addition of dithiothreitol
Phyre2
| 5 |
|
PDB 3cx5 chain C domain 2
Region: 3 - 47
Aligned: 45
Modelled: 45
Confidence: 7.4%
Identity: 7%
Fold: Heme-binding four-helical bundle
Superfamily: Transmembrane di-heme cytochromes
Family: Cytochrome b of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)
Phyre2
| 6 |
|
PDB 1m56 chain D
Region: 1 - 21
Aligned: 21
Modelled: 21
Confidence: 6.0%
Identity: 14%
Fold: Single transmembrane helix
Superfamily: Bacterial aa3 type cytochrome c oxidase subunit IV
Family: Bacterial aa3 type cytochrome c oxidase subunit IV
Phyre2
|
| Detailed template information | |
Due to computational demand, binding site predictions are not run for batch jobs
If you want to predict binding sites, please manually submit your model of choice to 3DLigandSite
Phyre is for academic use only
| Please cite: Protein structure prediction on
the web: a case study using the Phyre server |
| Kelley LA and Sternberg MJE. Nature Protocols
4, 363 - 371 (2009) [pdf] [Import into BibTeX] |
| |
| If you use the binding site
predictions from 3DLigandSite, please also cite: |
| 3DLigandSite: predicting ligand-binding sites using similar structures. |
| Wass MN, Kelley LA and Sternberg
MJ Nucleic Acids Research 38, W469-73 (2010) [PubMed] |
| |
|
|
|
|