| 1 |
|
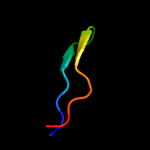
PDB 2jty chain A
Region: 20 - 46
Aligned: 27
Modelled: 27
Confidence: 35.9%
Identity: 15%
PDB header:structural protein
Chain: A: PDB Molecule:type-1 fimbrial protein, a chain;
PDBTitle: self-complemented variant of fima, the main subunit of type 1 pilus
Phyre2
| 2 |
|
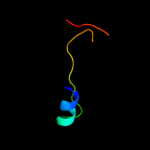
PDB 3jwn chain L
Region: 26 - 43
Aligned: 18
Modelled: 18
Confidence: 30.3%
Identity: 33%
PDB header:protein binding/cell adhesion
Chain: L: PDB Molecule:protein fimf;
PDBTitle: complex of fimc, fimf, fimg and fimh
Phyre2
| 3 |
|
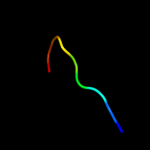
PDB 3jwn chain F
Region: 26 - 43
Aligned: 18
Modelled: 18
Confidence: 28.7%
Identity: 33%
PDB header:protein binding/cell adhesion
Chain: F: PDB Molecule:protein fimf;
PDBTitle: complex of fimc, fimf, fimg and fimh
Phyre2
| 4 |
|
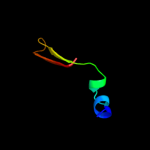
PDB 3jwn chain K
Region: 26 - 43
Aligned: 18
Modelled: 18
Confidence: 27.1%
Identity: 33%
PDB header:protein binding/cell adhesion
Chain: K: PDB Molecule:protein fimf;
PDBTitle: complex of fimc, fimf, fimg and fimh
Phyre2
| 5 |
|
PDB 3jwn chain E
Region: 26 - 43
Aligned: 18
Modelled: 18
Confidence: 27.1%
Identity: 33%
PDB header:protein binding/cell adhesion
Chain: E: PDB Molecule:protein fimf;
PDBTitle: complex of fimc, fimf, fimg and fimh
Phyre2
| 6 |
|
PDB 2jmr chain A
Region: 26 - 43
Aligned: 18
Modelled: 18
Confidence: 22.3%
Identity: 33%
PDB header:cell adhesion
Chain: A: PDB Molecule:fimf;
PDBTitle: nmr structure of the e. coli type 1 pilus subunit fimf
Phyre2
| 7 |
|
PDB 2j2z chain B domain 1
Region: 29 - 43
Aligned: 15
Modelled: 15
Confidence: 15.4%
Identity: 40%
Fold: Common fold of diphtheria toxin/transcription factors/cytochrome f
Superfamily: Bacterial adhesins
Family: Pilus subunits
Phyre2
| 8 |
|
PDB 3aq3 chain A
Region: 44 - 67
Aligned: 24
Modelled: 23
Confidence: 11.9%
Identity: 25%
PDB header:toxin
Chain: A: PDB Molecule:6b protein;
PDBTitle: molecular insights into plant cell proliferation disturbance by2 agrobacterium protein 6b
Phyre2
| 9 |
|
PDB 2eou chain A
Region: 50 - 61
Aligned: 12
Modelled: 12
Confidence: 10.3%
Identity: 25%
PDB header:transcription
Chain: A: PDB Molecule:zinc finger protein 473;
PDBTitle: solution structure of the c2h2 type zinc finger (region 370-2 400) of human zinc finger protein 473
Phyre2
| 10 |
|
PDB 2f09 chain A domain 1
Region: 24 - 41
Aligned: 18
Modelled: 18
Confidence: 8.4%
Identity: 28%
Fold: Streptavidin-like
Superfamily: YdhA-like
Family: YdhA-like
Phyre2
| 11 |
|
PDB 2pa2 chain A domain 1
Region: 43 - 70
Aligned: 28
Modelled: 28
Confidence: 8.2%
Identity: 18%
Fold: alpha/beta-Hammerhead
Superfamily: Ribosomal protein L16p/L10e
Family: Ribosomal protein L10e
Phyre2
| 12 |
|
PDB 2wkd chain A
Region: 19 - 34
Aligned: 16
Modelled: 16
Confidence: 8.0%
Identity: 31%
PDB header:dna binding protein
Chain: A: PDB Molecule:orf34p2;
PDBTitle: crystal structure of a double ile-to-met mutant of protein2 orf34 from lactococcus phage p2
Phyre2
| 13 |
|
PDB 2ctd chain A domain 1
Region: 32 - 40
Aligned: 9
Modelled: 9
Confidence: 7.7%
Identity: 56%
Fold: beta-beta-alpha zinc fingers
Superfamily: beta-beta-alpha zinc fingers
Family: Classic zinc finger, C2H2
Phyre2
| 14 |
|
PDB 2ob9 chain A
Region: 60 - 89
Aligned: 30
Modelled: 30
Confidence: 7.4%
Identity: 20%
PDB header:chaperone
Chain: A: PDB Molecule:tail assembly chaperone;
PDBTitle: structure of bacteriophage hk97 tail assembly chaperone
Phyre2