| 1 |
|
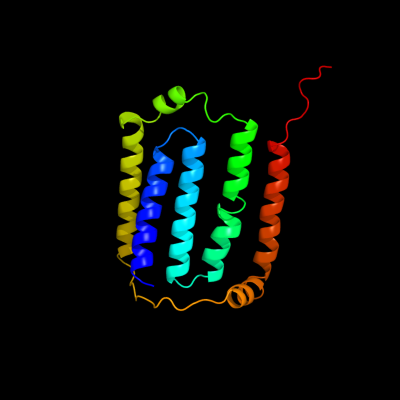
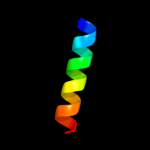
PDB 3rko chain F
Region: 1 - 168
Aligned: 168
Modelled: 168
Confidence: 100.0%
Identity: 100%
PDB header:oxidoreductase
Chain: F: PDB Molecule:nadh-quinone oxidoreductase subunit j;
PDBTitle: crystal structure of the membrane domain of respiratory complex i from2 e. coli at 3.0 angstrom resolution
Phyre2
| 2 |
|
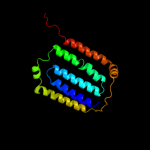
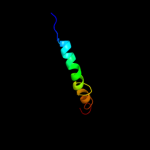
PDB 2fd5 chain A domain 2
Region: 145 - 178
Aligned: 33
Modelled: 34
Confidence: 67.9%
Identity: 21%
Fold: Tetracyclin repressor-like, C-terminal domain
Superfamily: Tetracyclin repressor-like, C-terminal domain
Family: Tetracyclin repressor-like, C-terminal domain
Phyre2
| 3 |
|
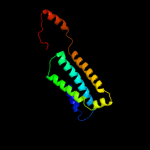
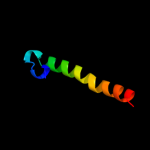
PDB 3kp9 chain A
Region: 4 - 138
Aligned: 130
Modelled: 135
Confidence: 20.4%
Identity: 15%
PDB header:blood coagulation,oxidoreductase
Chain: A: PDB Molecule:vkorc1/thioredoxin domain protein;
PDBTitle: structure of a bacterial homolog of vitamin k epoxide reductase
Phyre2
| 4 |
|
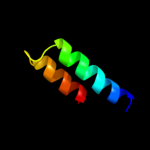
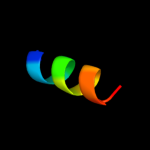
PDB 2a93 chain B
Region: 162 - 180
Aligned: 19
Modelled: 19
Confidence: 13.1%
Identity: 16%
PDB header:leucine zippers
Chain: B: PDB Molecule:c-myc-max heterodimeric leucine zipper;
PDBTitle: nmr solution structure of the c-myc-max heterodimeric2 leucine zipper, 40 structures
Phyre2
| 5 |
|
PDB 2qts chain A
Region: 23 - 60
Aligned: 36
Modelled: 38
Confidence: 12.4%
Identity: 28%
PDB header:membrane protein
Chain: A: PDB Molecule:acid-sensing ion channel;
PDBTitle: structure of an acid-sensing ion channel 1 at 1.9 a resolution and low2 ph
Phyre2
| 6 |
|
PDB 3b5o chain A
Region: 115 - 151
Aligned: 37
Modelled: 37
Confidence: 9.0%
Identity: 16%
PDB header:oxidoreductase
Chain: A: PDB Molecule:cadd-like protein of unknown function;
PDBTitle: crystal structure of a cadd-like protein of unknown function2 (npun_f6505) from nostoc punctiforme pcc 73102 at 1.35 a resolution
Phyre2
| 7 |
|
PDB 1h2s chain B
Region: 56 - 67
Aligned: 12
Modelled: 12
Confidence: 8.9%
Identity: 42%
Fold: Transmembrane helix hairpin
Superfamily: Htr2 transmembrane domain-like
Family: Htr2 transmembrane domain-like
Phyre2
| 8 |
|
PDB 1h2s chain B
Region: 56 - 67
Aligned: 12
Modelled: 12
Confidence: 8.9%
Identity: 42%
PDB header:membrane protein
Chain: B: PDB Molecule:sensory rhodopsin ii transducer;
PDBTitle: molecular basis of transmenbrane signalling by sensory2 rhodopsin ii-transducer complex
Phyre2
| 9 |
|
PDB 1u2m chain A
Region: 161 - 184
Aligned: 24
Modelled: 24
Confidence: 6.9%
Identity: 21%
Fold: OmpH-like
Superfamily: OmpH-like
Family: OmpH-like
Phyre2
| 10 |
|
PDB 2hep chain A
Region: 158 - 181
Aligned: 24
Modelled: 24
Confidence: 5.9%
Identity: 21%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:upf0291 protein ynzc;
PDBTitle: solution nmr structure of the upf0291 protein ynzc from2 bacillus subtilis. northeast structural genomics target3 sr384.
Phyre2
| 11 |
|
PDB 2hep chain A domain 1
Region: 158 - 181
Aligned: 24
Modelled: 24
Confidence: 5.9%
Identity: 21%
Fold: Long alpha-hairpin
Superfamily: YnzC-like
Family: YznC-like
Phyre2
| 12 |
|
PDB 2f95 chain B
Region: 58 - 67
Aligned: 10
Modelled: 10
Confidence: 5.4%
Identity: 50%
PDB header:membrane protein
Chain: B: PDB Molecule:sensory rhodopsin ii transducer;
PDBTitle: m intermediate structure of sensory rhodopsin ii/transducer complex in2 combination with the ground state structure
Phyre2
| 13 |
|
PDB 3rko chain K
Region: 2 - 90
Aligned: 89
Modelled: 89
Confidence: 5.4%
Identity: 13%
PDB header:oxidoreductase
Chain: K: PDB Molecule:nadh-quinone oxidoreductase subunit k;
PDBTitle: crystal structure of the membrane domain of respiratory complex i from2 e. coli at 3.0 angstrom resolution
Phyre2