| 1 |
|
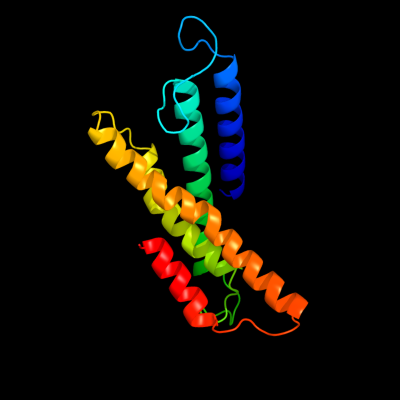
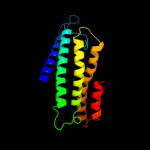
PDB 3dtu chain A domain 1
Region: 88 - 248
Aligned: 161
Modelled: 161
Confidence: 30.7%
Identity: 11%
Fold: Cytochrome c oxidase subunit I-like
Superfamily: Cytochrome c oxidase subunit I-like
Family: Cytochrome c oxidase subunit I-like
Phyre2
| 2 |
|
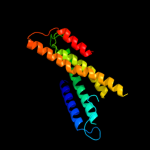
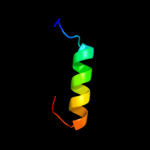
PDB 1m56 chain G
Region: 88 - 248
Aligned: 161
Modelled: 161
Confidence: 15.3%
Identity: 9%
PDB header:oxidoreductase
Chain: G: PDB Molecule:cytochrome c oxidase;
PDBTitle: structure of cytochrome c oxidase from rhodobactor2 sphaeroides (wild type)
Phyre2
| 3 |
|
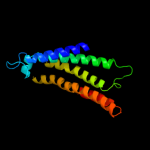
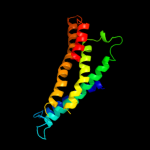
PDB 1v54 chain A
Region: 88 - 248
Aligned: 161
Modelled: 160
Confidence: 10.5%
Identity: 8%
Fold: Cytochrome c oxidase subunit I-like
Superfamily: Cytochrome c oxidase subunit I-like
Family: Cytochrome c oxidase subunit I-like
Phyre2
| 4 |
|
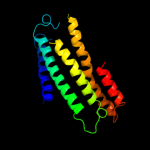
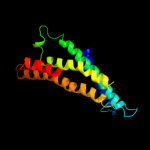
PDB 1ar1 chain A
Region: 88 - 248
Aligned: 161
Modelled: 161
Confidence: 9.6%
Identity: 13%
Fold: Cytochrome c oxidase subunit I-like
Superfamily: Cytochrome c oxidase subunit I-like
Family: Cytochrome c oxidase subunit I-like
Phyre2
| 5 |
|
PDB 3ol4 chain B
Region: 164 - 186
Aligned: 23
Modelled: 23
Confidence: 7.7%
Identity: 17%
PDB header:unknown function
Chain: B: PDB Molecule:putative uncharacterized protein;
PDBTitle: crystal structure of a putative uncharacterized protein from2 mycobacterium smegmatis, an ortholog of rv0543c
Phyre2
| 6 |
|
PDB 1fft chain F
Region: 88 - 248
Aligned: 161
Modelled: 161
Confidence: 5.9%
Identity: 10%
PDB header:oxidoreductase
Chain: F: PDB Molecule:ubiquinol oxidase;
PDBTitle: the structure of ubiquinol oxidase from escherichia coli
Phyre2
| 7 |
|
PDB 1fft chain A
Region: 88 - 248
Aligned: 161
Modelled: 161
Confidence: 5.9%
Identity: 10%
Fold: Cytochrome c oxidase subunit I-like
Superfamily: Cytochrome c oxidase subunit I-like
Family: Cytochrome c oxidase subunit I-like
Phyre2