| 1 | c3rpjA_ |
|
 |
100.0 |
48 |
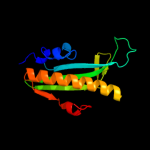
PDB header:transcription regulator
Chain: A: PDB Molecule:curlin genes transcriptional regulator;
PDBTitle: structure of a curlin genes transcriptional regulator protein from2 proteus mirabilis hi4320.
|
| 2 | d2bhua1 |
|
 |
67.4 |
20 |
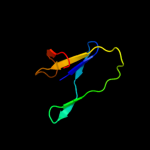
Fold:Immunoglobulin-like beta-sandwich
Superfamily:E set domains
Family:E-set domains of sugar-utilizing enzymes |
| 3 | c2b9bA_ |
|
 |
62.2 |
44 |
PDB header:viral protein
Chain: A: PDB Molecule:fusion glycoprotein f0;
PDBTitle: structure of the parainfluenza virus 5 f protein in its metastable,2 pre-fusion conformation
|
| 4 | d2a1va1 |
|
 |
49.9 |
18 |
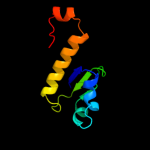
Fold:Secretion chaperone-like
Superfamily:YjbR-like
Family:YjbR-like |
| 5 | d3duea1 |
|
 |
45.9 |
23 |
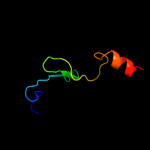
Fold:BLIP-like
Superfamily:BT0923-like
Family:BT0923-like |
| 6 | d2fkia1 |
|
 |
32.8 |
22 |
Fold:Secretion chaperone-like
Superfamily:YjbR-like
Family:YjbR-like |
| 7 | c3nrlB_ |
|
 |
22.2 |
25 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:uncharacterized protein rumgna_01417;
PDBTitle: crystal structure of protein rumgna_01417 from ruminococcus gnavus,2 northeast structural genomics consortium target ugr76
|
| 8 | d1axib1 |
|
 |
18.0 |
28 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Fibronectin type III
Family:Fibronectin type III |
| 9 | d3elga1 |
|
 |
17.7 |
15 |
Fold:BLIP-like
Superfamily:BT0923-like
Family:BT0923-like |
| 10 | c1agqB_ |
|
 |
16.4 |
31 |
PDB header:growth factor
Chain: B: PDB Molecule:glial cell-derived neurotrophic factor;
PDBTitle: glial cell-derived neurotrophic factor from rat
|
| 11 | d1b63a1 |
|
 |
16.0 |
21 |
Fold:Ribosomal protein S5 domain 2-like
Superfamily:Ribosomal protein S5 domain 2-like
Family:DNA gyrase/MutL, second domain |
| 12 | d2p02a2 |
|
 |
15.1 |
35 |
Fold:S-adenosylmethionine synthetase
Superfamily:S-adenosylmethionine synthetase
Family:S-adenosylmethionine synthetase |
| 13 | d1mxaa2 |
|
 |
14.3 |
30 |
Fold:S-adenosylmethionine synthetase
Superfamily:S-adenosylmethionine synthetase
Family:S-adenosylmethionine synthetase |
| 14 | d1qm4a2 |
|
 |
13.0 |
30 |
Fold:S-adenosylmethionine synthetase
Superfamily:S-adenosylmethionine synthetase
Family:S-adenosylmethionine synthetase |
| 15 | c2gyrB_ |
|
 |
11.9 |
13 |
PDB header:hormone/growth factor
Chain: B: PDB Molecule:neurotrophic factor artemin, isoform 3;
PDBTitle: crystal structure of human artemin
|
| 16 | d1zyma1 |
|
 |
10.4 |
19 |
Fold:SAM domain-like
Superfamily:Enzyme I of the PEP:sugar phosphotransferase system HPr-binding (sub)domain
Family:Enzyme I of the PEP:sugar phosphotransferase system HPr-binding (sub)domain |
| 17 | c2w82C_ |
|
 |
9.7 |
28 |
PDB header:replication inhibitor
Chain: C: PDB Molecule:orf18;
PDBTitle: the structure of arda
|
| 18 | c1lu0A_ |
|
 |
9.6 |
86 |
PDB header:hydrolase inhibitor
Chain: A: PDB Molecule:trypsin inhibitor i;
PDBTitle: atomic resolution structure of squash trypsin inhibitor: unexpected2 metal coordination
|
| 19 | d1lu0a_ |
|
 |
9.6 |
86 |
Fold:Knottins (small inhibitors, toxins, lectins)
Superfamily:Plant inhibitors of proteinases and amylases
Family:Plant inhibitors of proteinases and amylases |
| 20 | c3ctiA_ |
|
 |
9.6 |
86 |
PDB header:proteinase inhibitor (trypsin)
Chain: A: PDB Molecule:trypsin inhibitor;
PDBTitle: relaxation matrix refinement of the solution structure of2 squash trypsin inhibitor
|
| 21 | c2ctiA_ |
|
not modelled |
9.6 |
86 |
PDB header:proteinase inhibitor (trypsin)
Chain: A: PDB Molecule:trypsin inhibitor;
PDBTitle: determination of the complete three-dimensional structure2 of the trypsin inhibitor from squash seeds in aqueous3 solution by nuclear magnetic resonance and a combination4 of distance geometry and dynamical simulated annealing
|
| 22 | c1ppeI_ |
|
not modelled |
9.6 |
86 |
PDB header:hydrolase(serine proteinase)
Chain: I: PDB Molecule:trypsin inhibitor cmti-i;
PDBTitle: the refined 2.0 angstroms x-ray crystal structure of the2 complex formed between bovine beta-trypsin and cmti-i, a3 trypsin inhibitor from squash seeds (cucurbita maxima):4 topological similarity of the squash seed inhibitors with5 the carboxypeptidase a inhibitor from potatoes
|
| 23 | c1ctiA_ |
|
not modelled |
9.6 |
86 |
PDB header:proteinase inhibitor (trypsin)
Chain: A: PDB Molecule:trypsin inhibitor;
PDBTitle: determination of the complete three-dimensional structure2 of the trypsin inhibitor from squash seeds in aqueous3 solution by nuclear magnetic resonance and a combination4 of distance geometry and dynamical simulated annealing
|
| 24 | c2staI_ |
|
not modelled |
9.6 |
86 |
PDB header:hydrolase/hydrolase inhibitor
Chain: I: PDB Molecule:protein (trypsin inhibitor);
PDBTitle: anionic salmon trypsin in complex with squash seed2 inhibitor (cucurbita maxima trypsin inhibitor i)
|
| 25 | d2id1a1 |
|
not modelled |
9.5 |
25 |
Fold:Nucleotidyltransferase
Superfamily:Nucleotidyltransferase
Family:Iojap/YbeB-like |
| 26 | c1lu0B_ |
|
not modelled |
9.5 |
86 |
PDB header:hydrolase inhibitor
Chain: B: PDB Molecule:trypsin inhibitor i;
PDBTitle: atomic resolution structure of squash trypsin inhibitor: unexpected2 metal coordination
|
| 27 | c2v1vA_ |
|
not modelled |
9.5 |
86 |
PDB header:hydrolase inhibitor
Chain: A: PDB Molecule:trypsin inhibitor 1;
PDBTitle: 3d structure of the m8l mutant of squash trypsin inhibitor2 cmti-i
|
| 28 | c2a7oA_ |
|
not modelled |
9.2 |
50 |
PDB header:transcription
Chain: A: PDB Molecule:huntingtin interacting protein b;
PDBTitle: solution structure of the hset2/hypb sri domain
|
| 29 | d2c4ba2 |
|
not modelled |
9.2 |
86 |
Fold:Knottins (small inhibitors, toxins, lectins)
Superfamily:Plant inhibitors of proteinases and amylases
Family:Plant inhibitors of proteinases and amylases |
| 30 | c2vpzG_ |
|
not modelled |
9.2 |
50 |
PDB header:oxidoreductase
Chain: G: PDB Molecule:hypothetical membrane spanning protein;
PDBTitle: polysulfide reductase native structure
|
| 31 | d1h9ii_ |
|
not modelled |
9.1 |
60 |
Fold:Knottins (small inhibitors, toxins, lectins)
Superfamily:Plant inhibitors of proteinases and amylases
Family:Plant inhibitors of proteinases and amylases |
| 32 | d1h9hi_ |
|
not modelled |
8.8 |
86 |
Fold:Knottins (small inhibitors, toxins, lectins)
Superfamily:Plant inhibitors of proteinases and amylases
Family:Plant inhibitors of proteinases and amylases |
| 33 | c1mcvI_ |
|
not modelled |
8.8 |
86 |
PDB header:hydrolase
Chain: I: PDB Molecule:hei-toe i;
PDBTitle: crystal structure analysis of a hybrid squash inhibitor in2 complex with porcine pancreatic elastase
|
| 34 | d1mcvi_ |
|
not modelled |
8.8 |
86 |
Fold:Knottins (small inhibitors, toxins, lectins)
Superfamily:Plant inhibitors of proteinases and amylases
Family:Plant inhibitors of proteinases and amylases |
| 35 | c2it7A_ |
|
not modelled |
8.7 |
86 |
PDB header:plant protein
Chain: A: PDB Molecule:trypsin inhibitor 2;
PDBTitle: solution structure of the squash trypsin inhibitor eeti-ii
|
| 36 | d2it7a1 |
|
not modelled |
8.7 |
86 |
Fold:Knottins (small inhibitors, toxins, lectins)
Superfamily:Plant inhibitors of proteinases and amylases
Family:Plant inhibitors of proteinases and amylases |
| 37 | c2letA_ |
|
not modelled |
8.5 |
86 |
PDB header:proteinase inhibitor(trypsin)
Chain: A: PDB Molecule:trypsin inhibitor ii;
PDBTitle: an 1h nmr determination of the three dimensional structures2 of mirror image forms of a leu-5 variant of the trypsin3 inhibitor ecballium elaterium (eeti-ii)
|
| 38 | d1rpya_ |
|
not modelled |
8.1 |
19 |
Fold:SH2-like
Superfamily:SH2 domain
Family:SH2 domain |
| 39 | c3m07A_ |
|
not modelled |
8.0 |
11 |
PDB header:unknown function
Chain: A: PDB Molecule:putative alpha amylase;
PDBTitle: 1.4 angstrom resolution crystal structure of putative alpha2 amylase from salmonella typhimurium.
|
| 40 | c3fg7A_ |
|
not modelled |
7.6 |
16 |
PDB header:structural protein
Chain: A: PDB Molecule:villin-1;
PDBTitle: the crystal structure of villin domain 6
|
| 41 | d1xa6a2 |
|
not modelled |
7.6 |
18 |
Fold:SH2-like
Superfamily:SH2 domain
Family:SH2 domain |
| 42 | d2evra2 |
|
not modelled |
7.3 |
26 |
Fold:Cysteine proteinases
Superfamily:Cysteine proteinases
Family:NlpC/P60 |
| 43 | c1ik9C_ |
|
not modelled |
7.3 |
38 |
PDB header:gene regulation/ligase
Chain: C: PDB Molecule:dna ligase iv;
PDBTitle: crystal structure of a xrcc4-dna ligase iv complex
|
| 44 | d2o62a1 |
|
not modelled |
7.2 |
23 |
Fold:Lipocalins
Superfamily:Lipocalins
Family:All1756-like |
| 45 | d2i1sa1 |
|
not modelled |
6.9 |
27 |
Fold:MM3350-like
Superfamily:MM3350-like
Family:MM3350-like |
| 46 | d1exbe_ |
|
not modelled |
6.6 |
21 |
Fold:POZ domain
Superfamily:POZ domain
Family:Tetramerization domain of potassium channels |
| 47 | d2btci_ |
|
not modelled |
6.5 |
71 |
Fold:Knottins (small inhibitors, toxins, lectins)
Superfamily:Plant inhibitors of proteinases and amylases
Family:Plant inhibitors of proteinases and amylases |
| 48 | c2btcI_ |
|
not modelled |
6.5 |
71 |
PDB header:hydrolase/hydrolase inhibitor
Chain: I: PDB Molecule:protein (trypsin inhibitor);
PDBTitle: bovine trypsin in complex with squash seed inhibitor2 (cucurbita pepo trypsin inhibitor ii)
|
| 49 | c2stbI_ |
|
not modelled |
6.5 |
71 |
PDB header:hydrolase/hydrolase inhibitor
Chain: I: PDB Molecule:protein (trypsin inhibitor);
PDBTitle: anionic salmon trypsin in complex with squash seed2 inhibitor (cucurbita pepo trypsin inhibitor ii)
|
| 50 | c3h9xB_ |
|
not modelled |
6.3 |
13 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:uncharacterized protein pspto_3016;
PDBTitle: crystal structure of the pspto_3016 protein from2 pseudomonas syringae, northeast structural genomics3 consortium target psr293
|
| 51 | c2ysxA_ |
|
not modelled |
6.2 |
15 |
PDB header:signaling protein
Chain: A: PDB Molecule:signaling inositol polyphosphate phosphatase
PDBTitle: solution structure of the human ship sh2 domain
|
| 52 | c2yh5A_ |
|
not modelled |
6.1 |
12 |
PDB header:lipid binding protein
Chain: A: PDB Molecule:dapx protein;
PDBTitle: structure of the c-terminal domain of bamc
|
| 53 | c2xznZ_ |
|
not modelled |
6.1 |
11 |
PDB header:ribosome
Chain: Z: PDB Molecule:rps21e;
PDBTitle: crystal structure of the eukaryotic 40s ribosomal2 subunit in complex with initiation factor 1. this file3 contains the 40s subunit and initiation factor for4 molecule 2
|
| 54 | d1t6aa_ |
|
not modelled |
6.0 |
12 |
Fold:TBP-like
Superfamily:Rbstp2229 protein
Family:Rbstp2229 protein |
| 55 | d2eyva1 |
|
not modelled |
6.0 |
19 |
Fold:SH2-like
Superfamily:SH2 domain
Family:SH2 domain |
| 56 | d1eysl_ |
|
not modelled |
5.9 |
67 |
Fold:Bacterial photosystem II reaction centre, L and M subunits
Superfamily:Bacterial photosystem II reaction centre, L and M subunits
Family:Bacterial photosystem II reaction centre, L and M subunits |
| 57 | c2kkuA_ |
|
not modelled |
5.8 |
38 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: solution structure of protein af2351 from archaeoglobus2 fulgidus. northeast structural genomics consortium target3 att9/ontario center for structural proteomics target af2351
|
| 58 | d2bcqa3 |
|
not modelled |
5.5 |
25 |
Fold:Nucleotidyltransferase
Superfamily:Nucleotidyltransferase
Family:DNA polymerase beta-like |
| 59 | c2nvgA_ |
|
not modelled |
5.5 |
13 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:ubiquinol-cytochrome c reductase iron-sulfur subunit;
PDBTitle: soluble domain of rieske iron sulfur protein.
|
| 60 | c2qv5A_ |
|
not modelled |
5.5 |
23 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein atu2773;
PDBTitle: crystal structure of uncharacterized protein atu2773 from2 agrobacterium tumefaciens c58
|
| 61 | c3rv2B_ |
|
not modelled |
5.4 |
43 |
PDB header:transferase
Chain: B: PDB Molecule:s-adenosylmethionine synthase;
PDBTitle: crystal structure of s-adenosylmethionine synthetase from2 mycobacterium marinum
|

