| 1 |
|
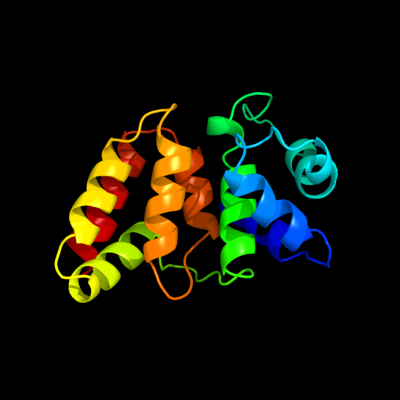
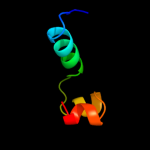
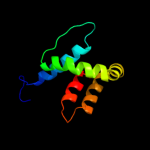
PDB 2zca chain B
Region: 6 - 156
Aligned: 147
Modelled: 151
Confidence: 100.0%
Identity: 16%
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:putative uncharacterized protein tthb189;
PDBTitle: crystal structure of tthb189, a crispr-associated protein,2 cse2 family from thermus thermophilus hb8
Phyre2
| 2 |
|
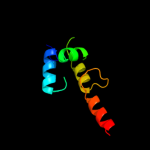
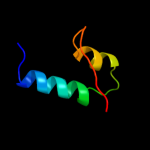
PDB 1k3k chain A
Region: 98 - 139
Aligned: 42
Modelled: 42
Confidence: 19.6%
Identity: 19%
Fold: Toxins' membrane translocation domains
Superfamily: Bcl-2 inhibitors of programmed cell death
Family: Bcl-2 inhibitors of programmed cell death
Phyre2
| 3 |
|
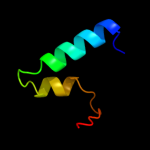
PDB 1shz chain F
Region: 103 - 155
Aligned: 51
Modelled: 53
Confidence: 9.1%
Identity: 16%
PDB header:signaling protein
Chain: F: PDB Molecule:rho guanine nucleotide exchange factor 1;
PDBTitle: crystal structure of the p115rhogef rgrgs domain in a2 complex with galpha(13):galpha(i1) chimera
Phyre2
| 4 |
|
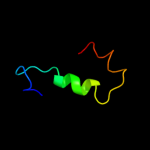
PDB 1h8b chain A
Region: 1 - 37
Aligned: 37
Modelled: 37
Confidence: 9.1%
Identity: 24%
Fold: EF Hand-like
Superfamily: EF-hand
Family: EF-hand modules in multidomain proteins
Phyre2
| 5 |
|
PDB 1j55 chain A
Region: 92 - 160
Aligned: 61
Modelled: 69
Confidence: 7.8%
Identity: 10%
Fold: EF Hand-like
Superfamily: EF-hand
Family: S100 proteins
Phyre2
| 6 |
|
PDB 1oh0 chain A
Region: 1 - 38
Aligned: 38
Modelled: 38
Confidence: 7.1%
Identity: 16%
Fold: Cystatin-like
Superfamily: NTF2-like
Family: Ketosteroid isomerase-like
Phyre2
| 7 |
|
PDB 1ysy chain A domain 1
Region: 105 - 138
Aligned: 34
Modelled: 34
Confidence: 6.6%
Identity: 15%
Fold: immunoglobulin/albumin-binding domain-like
Superfamily: Coronavirus NSP7-like
Family: Coronavirus NSP7-like
Phyre2
| 8 |
|
PDB 2kua chain A
Region: 46 - 139
Aligned: 94
Modelled: 94
Confidence: 6.4%
Identity: 4%
PDB header:apoptosis
Chain: A: PDB Molecule:bcl-2-like protein 10;
PDBTitle: solution structure of a divergent bcl-2 protein
Phyre2
| 9 |
|
PDB 1ohp chain A domain 1
Region: 1 - 38
Aligned: 38
Modelled: 38
Confidence: 6.1%
Identity: 18%
Fold: Cystatin-like
Superfamily: NTF2-like
Family: Ketosteroid isomerase-like
Phyre2