| 1 |
|
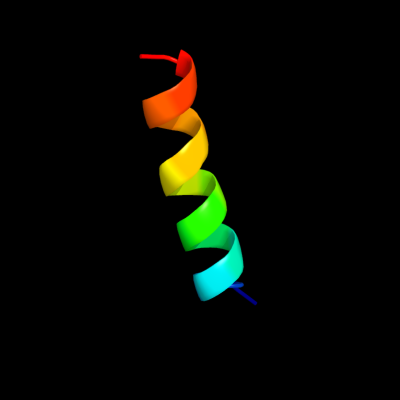
PDB 1q90 chain G
Region: 1 - 19
Aligned: 19
Modelled: 19
Confidence: 25.9%
Identity: 37%
PDB header:photosynthesis
Chain: G: PDB Molecule:cytochrome b6f complex subunit petg;
PDBTitle: structure of the cytochrome b6f (plastohydroquinone : plastocyanin2 oxidoreductase) from chlamydomonas reinhardtii
Phyre2
| 2 |
|
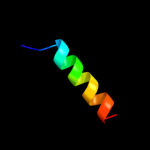
PDB 1q90 chain G
Region: 1 - 19
Aligned: 19
Modelled: 19
Confidence: 25.9%
Identity: 37%
Fold: Single transmembrane helix
Superfamily: PetG subunit of the cytochrome b6f complex
Family: PetG subunit of the cytochrome b6f complex
Phyre2
| 3 |
|
PDB 2hjm chain B
Region: 5 - 37
Aligned: 33
Modelled: 33
Confidence: 13.1%
Identity: 24%
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:hypothetical protein pf1176;
PDBTitle: crystal structure of a singleton protein pf1176 from p. furiosus
Phyre2
| 4 |
|
PDB 2kqz chain A
Region: 4 - 21
Aligned: 18
Modelled: 18
Confidence: 5.8%
Identity: 33%
PDB header:protein binding
Chain: A: PDB Molecule:proteasomal ubiquitin receptor adrm1;
PDBTitle: fragment of proteasome protein
Phyre2
| 5 |
|
PDB 2koe chain A
Region: 4 - 19
Aligned: 16
Modelled: 16
Confidence: 5.7%
Identity: 19%
PDB header:membrane protein, signaling protein
Chain: A: PDB Molecule:human cannabinoid receptor 1 - helix 7/8 peptide;
PDBTitle: human cannabinoid receptor 1 - helix 7/8 peptide
Phyre2
| 6 |
|
PDB 1xnl chain A
Region: 6 - 23
Aligned: 18
Modelled: 18
Confidence: 5.5%
Identity: 28%
PDB header:viral protein
Chain: A: PDB Molecule:membrane protein gp37;
PDBTitle: aslv fusion peptide
Phyre2
| 7 |
|
PDB 3bya chain B
Region: 2 - 16
Aligned: 15
Modelled: 15
Confidence: 5.4%
Identity: 33%
PDB header:metal binding protein
Chain: B: PDB Molecule:glutamate [nmda] receptor subunit zeta-1 peptide;
PDBTitle: structure of a calmodulin complex
Phyre2