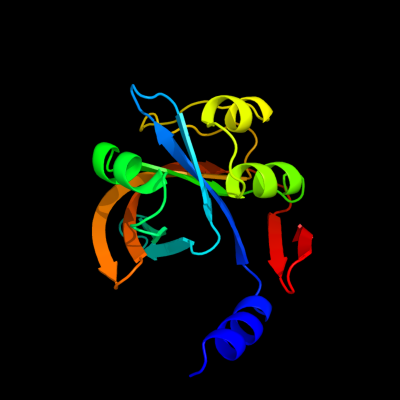
| 1 | c2r0xA_
|
|
 |
100.0 |
50 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:possible flavin reductase;
PDBTitle: crystal structure of a putative flavin reductase (ycdh, hs_1225) from2 haemophilus somnus 129pt at 1.06 a resolution
|
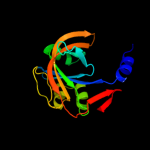
| 2 | c3k87B_
|
|
 |
100.0 |
41 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:chlorophenol-4-monooxygenase component 1;
PDBTitle: crystal structure of nadh:fad oxidoreductase (tftc) - fad2 complex
|
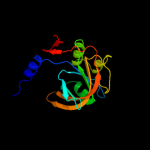
| 3 | c3rh7A_
|
|
 |
100.0 |
31 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:hypothetical oxidoreductase;
PDBTitle: crystal structure of a hypothetical oxidoreductase (sma0793) from2 sinorhizobium meliloti 1021 at 3.00 a resolution
|
| 4 | c3cb0B_
|
|
 |
100.0 |
39 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:4-hydroxyphenylacetate 3-monooxygenase;
PDBTitle: cobr
|
| 5 | c3nfwB_
|
|
 |
100.0 |
25 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:nitrilotriacetate monooxygenase component b;
PDBTitle: crystal structure of nitrilotriacetate monooxygenase component b2 (a0r521 homolog) from mycobacterium thermoresistibile
|
| 6 | c3pftA_
|
|
 |
100.0 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:flavin reductase;
PDBTitle: crystal structure of untagged c54a mutant flavin reductase (dszd) in2 complex with fmn from mycobacterium goodii
|
| 7 | c2qckA_
|
|
 |
100.0 |
22 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:flavin reductase domain protein;
PDBTitle: crystal structure of flavin reductase domain protein (yp_831077.1)2 from arthrobacter sp. fb24 at 1.90 a resolution
|
| 8 | c2d38A_
|
|
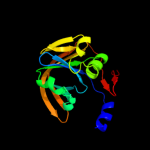 |
100.0 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:hypothetical nadh-dependent fmn oxidoreductase;
PDBTitle: the crystal structure of flavin reductase hpac complexed with nadp+
|
| 9 | c2ecrA_
|
|
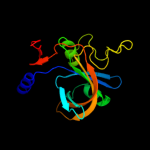 |
100.0 |
26 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:flavin reductase component (hpac) of 4-hydroxyphenylacetate
PDBTitle: crystal structure of the ligand-free form of the flavin reductase2 component (hpac) of 4-hydroxyphenylacetate 3-monooxygenase
|
| 10 | d1wgba_
|
|
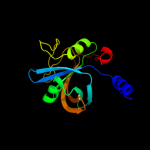 |
100.0 |
18 |
Fold:Split barrel-like
Superfamily:FMN-binding split barrel
Family:NADH:FMN oxidoreductase-like |
| 11 | d1rz0a_
|
|
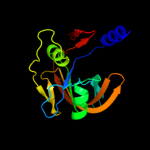 |
100.0 |
27 |
Fold:Split barrel-like
Superfamily:FMN-binding split barrel
Family:NADH:FMN oxidoreductase-like |
| 12 | d1i0ra_
|
|
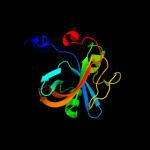 |
100.0 |
21 |
Fold:Split barrel-like
Superfamily:FMN-binding split barrel
Family:NADH:FMN oxidoreductase-like |
| 13 | c3bnkB_
|
|
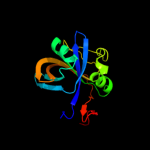 |
100.0 |
18 |
PDB header:electron transport
Chain: B: PDB Molecule:flavoredoxin;
PDBTitle: x-ray crystal structure of flavoredoxin from methanosarcina2 acetivorans
|
| 14 | c2r6vA_
|
|
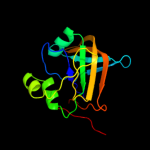 |
100.0 |
14 |
PDB header:flavoprotein
Chain: A: PDB Molecule:uncharacterized protein ph0856;
PDBTitle: crystal structure of fmn-binding protein (np_142786.1) from pyrococcus2 horikoshii at 1.35 a resolution
|
| 15 | d1ejea_
|
|
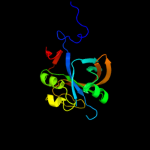 |
100.0 |
13 |
Fold:Split barrel-like
Superfamily:FMN-binding split barrel
Family:NADH:FMN oxidoreductase-like |
| 16 | d1usca_
|
|
 |
100.0 |
16 |
Fold:Split barrel-like
Superfamily:FMN-binding split barrel
Family:NADH:FMN oxidoreductase-like |
| 17 | c3bpkB_
|
|
 |
100.0 |
17 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:nitrilotriacetate monooxygenase component b;
PDBTitle: crystal structure of nitrilotriacetate monooxygenase2 component b from bacillus cereus
|
| 18 | c3e4vA_
|
|
 |
100.0 |
12 |
PDB header:flavoprotein
Chain: A: PDB Molecule:nadh:fmn oxidoreductase like protein;
PDBTitle: crystal structure of nadh:fmn oxidoreductase like protein in complex2 with fmn (yp_544701.1) from methylobacillus flagellatus kt at 1.40 a3 resolution
|
| 19 | c3hmzA_
|
|
 |
100.0 |
11 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:flavin reductase domain protein, fmn-binding;
PDBTitle: crystal structure of a fmn-binding domain of flavin reductases-like2 enzyme (sbal_0626) from shewanella baltica os155 at 1.50 a resolution
|
| 20 | c2d5mA_
|
|
 |
100.0 |
13 |
PDB header:electron transport
Chain: A: PDB Molecule:flavoredoxin;
PDBTitle: flavoredoxin of desulfovibrio vulgaris (miyazaki f)
|
| 21 | c3fgeA_ |
|
not modelled |
100.0 |
11 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:putative flavin reductase with split barrel domain;
PDBTitle: crystal structure of putative flavin reductase with split barrel2 domain (yp_750721.1) from shewanella frigidimarina ncimb 400 at 1.743 a resolution
|
| 22 | c3b5mD_ |
|
not modelled |
99.9 |
13 |
PDB header:structural genomics, unknown function
Chain: D: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of conserved uncharacterized protein from2 rhodopirellula baltica
|
| 23 | d2imla1 |
|
not modelled |
99.3 |
11 |
Fold:Split barrel-like
Superfamily:FMN-binding split barrel
Family:MTH863-like |
| 24 | c2ptfB_ |
|
not modelled |
98.6 |
14 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:uncharacterized protein mth_863;
PDBTitle: crystal structure of protein mth_863 from methanobacterium2 thermoautotrophicum bound to fmn
|
| 25 | d2nr4a1 |
|
not modelled |
98.6 |
16 |
Fold:Split barrel-like
Superfamily:FMN-binding split barrel
Family:MTH863-like |
| 26 | d2ptfa1 |
|
not modelled |
98.5 |
15 |
Fold:Split barrel-like
Superfamily:FMN-binding split barrel
Family:MTH863-like |
| 27 | d1flma_ |
|
not modelled |
95.3 |
12 |
Fold:Split barrel-like
Superfamily:FMN-binding split barrel
Family:PNP-oxidase like |
| 28 | c2q9kA_ |
|
not modelled |
95.0 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of a putative oxidoreductase (exig_1997) from2 exiguobacterium sibiricum 255-15 at 1.59 a resolution
|
| 29 | c2htdB_ |
|
not modelled |
88.3 |
6 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:predicted flavin-nucleotide-binding protein from cog3576
PDBTitle: crystal structure of a putative pyridoxamine 5'-phosphate oxidase2 (ldb0262) from lactobacillus delbrueckii subsp. at 1.60 a resolution
|
| 30 | d1vl7a_ |
|
not modelled |
78.9 |
18 |
Fold:Split barrel-like
Superfamily:FMN-binding split barrel
Family:PNP-oxidase like |
| 31 | d2hq7a1 |
|
not modelled |
70.0 |
10 |
Fold:Split barrel-like
Superfamily:FMN-binding split barrel
Family:PNP-oxidase like |
| 32 | c2re7A_ |
|
not modelled |
60.5 |
7 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of a pyridoxamine 5'-phosphate oxidase related2 protein (psyc_0186) from psychrobacter arcticus 273-4 at 2.50 a3 resolution
|
| 33 | d1rfea_ |
|
not modelled |
60.2 |
12 |
Fold:Split barrel-like
Superfamily:FMN-binding split barrel
Family:PNP-oxidase like |
| 34 | d2asfa1 |
|
not modelled |
54.1 |
8 |
Fold:Split barrel-like
Superfamily:FMN-binding split barrel
Family:PNP-oxidase like |
| 35 | d2htia1 |
|
not modelled |
51.9 |
8 |
Fold:Split barrel-like
Superfamily:FMN-binding split barrel
Family:PNP-oxidase like |
| 36 | c2htiA_ |
|
not modelled |
51.9 |
8 |
PDB header:fmn-binding protein
Chain: A: PDB Molecule:bh0577 protein;
PDBTitle: crystal structure of a flavin-nucleotide-binding protein (bh_0577)2 from bacillus halodurans at 2.50 a resolution
|
| 37 | c3ec6A_ |
|
not modelled |
46.4 |
11 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:general stress protein 26;
PDBTitle: crystal structure of the general stress protein 26 from bacillus2 anthracis str. sterne
|
| 38 | d2fhqa1 |
|
not modelled |
46.4 |
9 |
Fold:Split barrel-like
Superfamily:FMN-binding split barrel
Family:PNP-oxidase like |
| 39 | c3dmbA_ |
|
not modelled |
43.2 |
11 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:putative general stress protein 26 with a pnp-oxidase like
PDBTitle: crystal structure of a putative general stress family protein2 (xcc2264) from xanthomonas campestris pv. campestris at 2.30 a3 resolution
|
| 40 | c3db0B_ |
|
not modelled |
36.1 |
11 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:lin2891 protein;
PDBTitle: crystal structure of putative pyridoxamine 5'-phosphate oxidase2 (np_472219.1) from listeria innocua at 2.00 a resolution
|
| 41 | c3f7eB_ |
|
not modelled |
34.6 |
10 |
PDB header:unknown function
Chain: B: PDB Molecule:pyridoxamine 5'-phosphate oxidase-related, fmn-
PDBTitle: msmeg_3380 f420 reductase
|
| 42 | d1w9aa_ |
|
not modelled |
24.7 |
8 |
Fold:Split barrel-like
Superfamily:FMN-binding split barrel
Family:PNP-oxidase like |
| 43 | c3fkhB_ |
|
not modelled |
22.5 |
11 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:putative pyridoxamine 5'-phosphate oxidase;
PDBTitle: crystal structure of putative pyridoxamine 5'-phosphate oxidase2 (np_601736.1) from corynebacterium glutamicum atcc 13032 kitasato at3 2.51 a resolution
|
| 44 | d1xzwa1 |
|
not modelled |
19.0 |
18 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Purple acid phosphatase, N-terminal domain
Family:Purple acid phosphatase, N-terminal domain |
| 45 | d2arza1 |
|
not modelled |
17.4 |
15 |
Fold:Split barrel-like
Superfamily:FMN-binding split barrel
Family:PNP-oxidase like |
| 46 | d2hq9a1 |
|
not modelled |
16.2 |
6 |
Fold:Split barrel-like
Superfamily:FMN-binding split barrel
Family:PNP-oxidase like |
| 47 | c2ou5B_ |
|
not modelled |
11.6 |
12 |
PDB header:flavoprotein
Chain: B: PDB Molecule:pyridoxamine 5'-phosphate oxidase-related, fmn-binding;
PDBTitle: crystal structure of a pyridoxamine 5'-phosphate oxidase-related fmn-2 binding protein (jann_0254) from jannaschia sp. ccs1 at 1.60 a3 resolution
|
| 48 | c2ig6B_ |
|
not modelled |
9.5 |
13 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:nimc/nima family protein;
PDBTitle: crystal structure of a nimc/nima family protein (ca_c2569) from2 clostridium acetobutylicum at 1.80 a resolution
|
| 49 | d2qfra1 |
|
not modelled |
9.2 |
11 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Purple acid phosphatase, N-terminal domain
Family:Purple acid phosphatase, N-terminal domain |
| 50 | c2iabB_ |
|
not modelled |
8.1 |
14 |
PDB header:unknown function
Chain: B: PDB Molecule:hypothetical protein;
PDBTitle: crystal structure of a protein with fmn-binding split barrel fold2 (np_828636.1) from streptomyces avermitilis at 2.00 a resolution
|
| 51 | d2i02a1 |
|
not modelled |
7.9 |
13 |
Fold:Split barrel-like
Superfamily:FMN-binding split barrel
Family:PNP-oxidase like |
| 52 | d1mkya3 |
|
not modelled |
7.3 |
25 |
Fold:Alpha-lytic protease prodomain-like
Superfamily:Probable GTPase Der, C-terminal domain
Family:Probable GTPase Der, C-terminal domain |
| 53 | c3cp3A_ |
|
not modelled |
7.3 |
13 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of conserved protein of unknown function dip18742 from corynebacterium diphtheriae
|
| 54 | c1a2oB_ |
|
not modelled |
7.1 |
17 |
PDB header:bacterial chemotaxis
Chain: B: PDB Molecule:cheb methylesterase;
PDBTitle: structural basis for methylesterase cheb regulation by a2 phosphorylation-activated domain
|
| 55 | d1dmla2 |
|
not modelled |
7.0 |
24 |
Fold:DNA clamp
Superfamily:DNA clamp
Family:DNA polymerase processivity factor |
| 56 | d2fg9a1 |
|
not modelled |
6.3 |
15 |
Fold:Split barrel-like
Superfamily:FMN-binding split barrel
Family:PNP-oxidase like |
| 57 | c2hhzA_ |
|
not modelled |
6.0 |
6 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:pyridoxamine 5'-phosphate oxidase-related;
PDBTitle: crystal structure of a pyridoxamine 5'-phosphate oxidase-related2 protein (ssuidraft_2804) from streptococcus suis 89/1591 at 2.00 a3 resolution
|
| 58 | d1svba1 |
|
not modelled |
5.9 |
13 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:E set domains
Family:Class II viral fusion proteins C-terminal domain |
| 59 | c1mkyA_ |
|
not modelled |
5.4 |
20 |
PDB header:ligand binding protein
Chain: A: PDB Molecule:probable gtp-binding protein enga;
PDBTitle: structural analysis of the domain interactions in der, a2 switch protein containing two gtpase domains
|