| 1 |
|
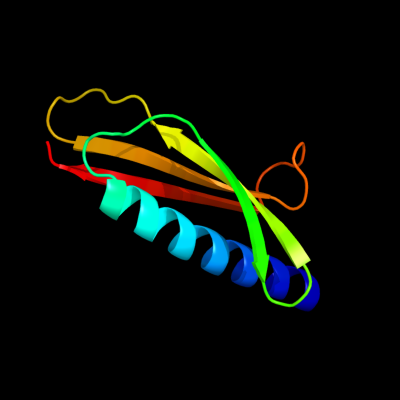
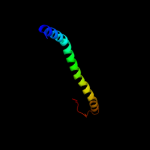
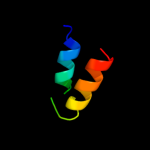
PDB 2ret chain E
Region: 36 - 117
Aligned: 82
Modelled: 82
Confidence: 99.7%
Identity: 17%
PDB header:protein transport
Chain: E: PDB Molecule:pseudopilin epsi;
PDBTitle: the crystal structure of a binary complex of two pseudopilins: epsi2 and epsj from the type 2 secretion system of vibrio vulnificus
Phyre2
| 2 |
|
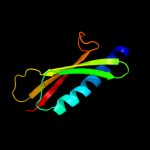
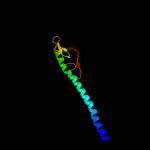
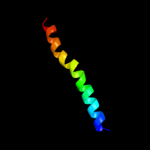
PDB 2ret chain A domain 1
Region: 36 - 115
Aligned: 80
Modelled: 80
Confidence: 99.7%
Identity: 18%
Fold: Pili subunits
Superfamily: Pili subunits
Family: GSPII I/J protein-like
Phyre2
| 3 |
|
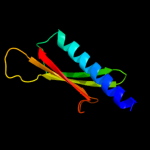
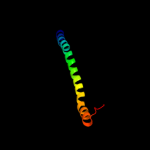
PDB 3ci0 chain I domain 1
Region: 38 - 115
Aligned: 77
Modelled: 78
Confidence: 99.6%
Identity: 21%
Fold: Pili subunits
Superfamily: Pili subunits
Family: GSPII I/J protein-like
Phyre2
| 4 |
|
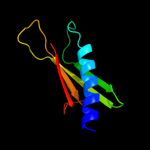
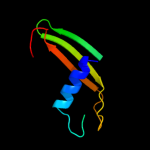
PDB 3sok chain B
Region: 7 - 69
Aligned: 63
Modelled: 63
Confidence: 98.1%
Identity: 14%
PDB header:cell adhesion
Chain: B: PDB Molecule:fimbrial protein;
PDBTitle: dichelobacter nodosus pilin fima
Phyre2
| 5 |
|
PDB 1oqw chain A
Region: 7 - 112
Aligned: 102
Modelled: 106
Confidence: 98.1%
Identity: 12%
Fold: Pili subunits
Superfamily: Pili subunits
Family: Pilin
Phyre2
| 6 |
|
PDB 2pil chain A
Region: 8 - 66
Aligned: 59
Modelled: 59
Confidence: 97.5%
Identity: 15%
Fold: Pili subunits
Superfamily: Pili subunits
Family: Pilin
Phyre2
| 7 |
|
PDB 4a18 chain U
Region: 5 - 15
Aligned: 11
Modelled: 11
Confidence: 24.9%
Identity: 45%
PDB header:ribosome
Chain: U: PDB Molecule:rpl13;
PDBTitle: t.thermophila 60s ribosomal subunit in complex with initiation2 factor 6. this file contains 26s rrna and proteins of molecule 1
Phyre2
| 8 |
|
PDB 3u5e chain L
Region: 5 - 15
Aligned: 11
Modelled: 11
Confidence: 23.5%
Identity: 55%
PDB header:ribosome
Chain: L: PDB Molecule:60s ribosomal protein l13-a;
PDBTitle: the structure of the eukaryotic ribosome at 3.0 resolution
Phyre2
| 9 |
|
PDB 2wsf chain G
Region: 2 - 13
Aligned: 12
Modelled: 12
Confidence: 20.6%
Identity: 33%
PDB header:photosynthesis
Chain: G: PDB Molecule:photosystem i reaction center subunit v,
PDBTitle: improved model of plant photosystem i
Phyre2
| 10 |
|
PDB 3lw5 chain K
Region: 2 - 13
Aligned: 12
Modelled: 12
Confidence: 16.6%
Identity: 33%
PDB header:photosynthesis
Chain: K: PDB Molecule:photosystem i reaction center subunit x psak;
PDBTitle: improved model of plant photosystem i
Phyre2
| 11 |
|
PDB 2o01 chain G
Region: 6 - 13
Aligned: 8
Modelled: 8
Confidence: 13.5%
Identity: 38%
PDB header:photosynthesis
Chain: G: PDB Molecule:photosystem i reaction center subunit v,
PDBTitle: the structure of a plant photosystem i supercomplex at 3.42 angstrom resolution
Phyre2
| 12 |
|
PDB 2wsc chain K
Region: 2 - 13
Aligned: 12
Modelled: 12
Confidence: 11.6%
Identity: 33%
PDB header:photosynthesis
Chain: K: PDB Molecule:photosystem i reaction center subunit psak,
PDBTitle: improved model of plant photosystem i
Phyre2
| 13 |
|
PDB 2axt chain H domain 1
Region: 14 - 34
Aligned: 21
Modelled: 21
Confidence: 10.0%
Identity: 14%
Fold: Single transmembrane helix
Superfamily: Photosystem II 10 kDa phosphoprotein PsbH
Family: PsbH-like
Phyre2
| 14 |
|
PDB 2k3p chain A
Region: 9 - 17
Aligned: 9
Modelled: 9
Confidence: 7.5%
Identity: 56%
PDB header:structural protein
Chain: A: PDB Molecule:tusp1;
PDBTitle: solution structure of the c-terminal domain (tusp1-c) of the2 egg case silk from nephila antipodiana
Phyre2
| 15 |
|
PDB 2bo9 chain B domain 1
Region: 44 - 108
Aligned: 65
Modelled: 65
Confidence: 7.3%
Identity: 15%
Fold: Cystatin-like
Superfamily: Cystatin/monellin
Family: Latexin-like
Phyre2
| 16 |
|
PDB 1p58 chain F
Region: 3 - 30
Aligned: 28
Modelled: 28
Confidence: 7.2%
Identity: 21%
PDB header:virus
Chain: F: PDB Molecule:envelope protein m;
PDBTitle: complex organization of dengue virus membrane proteins as revealed by2 9.5 angstrom cryo-em reconstruction
Phyre2
| 17 |
|
PDB 2ko6 chain A
Region: 3 - 17
Aligned: 15
Modelled: 15
Confidence: 6.9%
Identity: 27%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein yihd;
PDBTitle: solution structure of protein sf3929 from shigella flexneri2 2a. northeast structural genomics consortium target3 sfr81/ontario center for structural proteomics target4 sf3929
Phyre2
| 18 |
|
PDB 2kb1 chain A
Region: 7 - 38
Aligned: 32
Modelled: 32
Confidence: 6.4%
Identity: 9%
PDB header:membrane protein
Chain: A: PDB Molecule:wsk3;
PDBTitle: nmr studies of a channel protein without membrane:2 structure and dynamics of water-solubilized kcsa
Phyre2