| 1 |
|
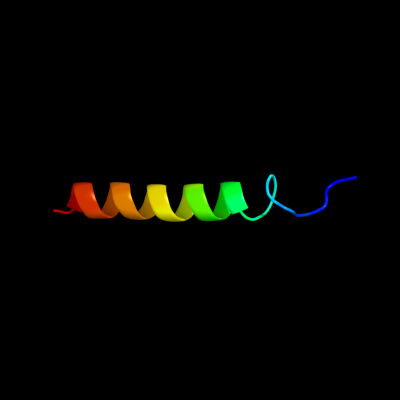
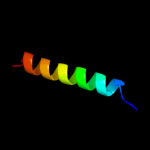
PDB 1bhb chain A
Region: 39 - 66
Aligned: 27
Modelled: 28
Confidence: 18.9%
Identity: 37%
PDB header:photoreceptor
Chain: A: PDB Molecule:bacteriorhodopsin;
PDBTitle: three-dimensional structure of (1-71) bacterioopsin2 solubilized in methanol-chloroform and sds micelles3 determined by 15n-1h heteronuclear nmr spectroscopy
Phyre2
| 2 |
|
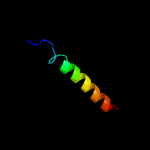
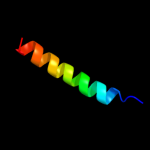
PDB 2kv5 chain A
Region: 50 - 80
Aligned: 26
Modelled: 31
Confidence: 18.0%
Identity: 23%
PDB header:toxin
Chain: A: PDB Molecule:putative uncharacterized protein rnai;
PDBTitle: solution structure of the par toxin fst in dpc micelles
Phyre2
| 3 |
|
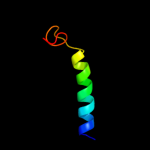
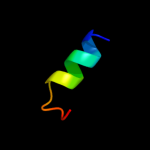
PDB 2knc chain B
Region: 43 - 72
Aligned: 29
Modelled: 30
Confidence: 14.3%
Identity: 17%
PDB header:cell adhesion
Chain: B: PDB Molecule:integrin beta-3;
PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
Phyre2
| 4 |
|
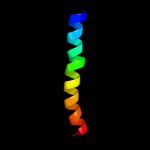
PDB 2rmz chain A
Region: 46 - 72
Aligned: 26
Modelled: 27
Confidence: 10.9%
Identity: 19%
PDB header:cell adhesion
Chain: A: PDB Molecule:integrin beta-3;
PDBTitle: bicelle-embedded integrin beta3 transmembrane segment
Phyre2
| 5 |
|
PDB 2k1a chain A
Region: 41 - 65
Aligned: 25
Modelled: 25
Confidence: 10.5%
Identity: 20%
PDB header:cell adhesion
Chain: A: PDB Molecule:integrin alpha-iib;
PDBTitle: bicelle-embedded integrin alpha(iib) transmembrane segment
Phyre2
| 6 |
|
PDB 2knc chain A
Region: 41 - 65
Aligned: 25
Modelled: 25
Confidence: 10.1%
Identity: 20%
PDB header:cell adhesion
Chain: A: PDB Molecule:integrin alpha-iib;
PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
Phyre2
| 7 |
|
PDB 3c1i chain A
Region: 4 - 47
Aligned: 44
Modelled: 44
Confidence: 8.7%
Identity: 20%
PDB header:transport protein
Chain: A: PDB Molecule:ammonia channel;
PDBTitle: substrate binding, deprotonation and selectivity at the2 periplasmic entrance of the e. coli ammonia channel amtb
Phyre2
| 8 |
|
PDB 1h5o chain A
Region: 5 - 11
Aligned: 7
Modelled: 7
Confidence: 5.9%
Identity: 43%
Fold: Defensin-like
Superfamily: Defensin-like
Family: Myotoxin
Phyre2
| 9 |
|
PDB 2d46 chain A
Region: 71 - 80
Aligned: 10
Modelled: 10
Confidence: 5.8%
Identity: 50%
PDB header:metal transport
Chain: A: PDB Molecule:calcium channel, voltage-dependent, beta 4
PDBTitle: solution structure of the human beta4a-a domain
Phyre2
| 10 |
|
PDB 1ehs chain A
Region: 1 - 11
Aligned: 11
Modelled: 11
Confidence: 5.6%
Identity: 27%
Fold: Toxic hairpin
Superfamily: Heat-stable enterotoxin B
Family: Heat-stable enterotoxin B
Phyre2
| 11 |
|
PDB 1ufi chain D
Region: 62 - 76
Aligned: 15
Modelled: 15
Confidence: 5.5%
Identity: 40%
PDB header:dna binding protein
Chain: D: PDB Molecule:major centromere autoantigen b;
PDBTitle: crystal structure of the dimerization domain of human cenp-b
Phyre2
| 12 |
|
PDB 1ufi chain A
Region: 62 - 76
Aligned: 15
Modelled: 15
Confidence: 5.2%
Identity: 40%
Fold: ROP-like
Superfamily: Dimerisation domain of CENP-B
Family: Dimerisation domain of CENP-B
Phyre2