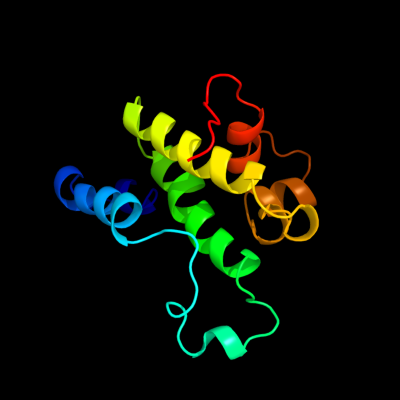
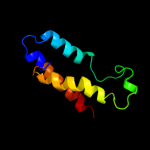
1 d1n7oa1
69.9
11
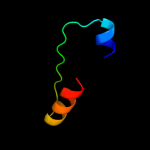
Fold: alpha/alpha toroidSuperfamily: Chondroitin AC/alginate lyaseFamily: Hyaluronate lyase-like catalytic, N-terminal domain2 d2d0ta1
49.8
12
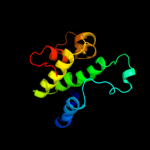
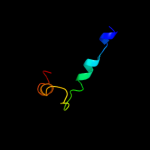
Fold: Indolic compounds 2,3-dioxygenase-likeSuperfamily: Indolic compounds 2,3-dioxygenase-likeFamily: Indoleamine 2,3-dioxygenase-like3 d1kyfa1
33.3
29
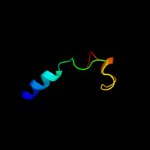
Fold: Immunoglobulin-like beta-sandwichSuperfamily: Clathrin adaptor appendage domainFamily: Alpha-adaptin ear subdomain-like4 d1rwha1
32.2
10
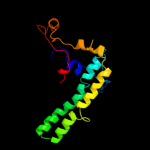
Fold: alpha/alpha toroidSuperfamily: Chondroitin AC/alginate lyaseFamily: Hyaluronate lyase-like catalytic, N-terminal domain5 c1qysA_
26.2
19
PDB header: de novo proteinChain: A: PDB Molecule: top7;PDBTitle: crystal structure of top7: a computationally designed2 protein with a novel fold
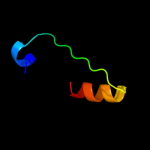
6 d1f1sa1
23.7
5
Fold: alpha/alpha toroidSuperfamily: Chondroitin AC/alginate lyaseFamily: Hyaluronate lyase-like catalytic, N-terminal domain7 c2jvfA_
19.1
26
PDB header: de novo proteinChain: A: PDB Molecule: de novo protein m7;PDBTitle: solution structure of m7, a computationally-designed2 artificial protein
8 d1e85a_
19.0
3
Fold: Four-helical up-and-down bundleSuperfamily: CytochromesFamily: Cytochrome c'-like9 d1mqva_
18.5
20
Fold: Four-helical up-and-down bundleSuperfamily: CytochromesFamily: Cytochrome c'-like10 d2nr7a1
18.3
20
Fold: Lysozyme-likeSuperfamily: Lysozyme-likeFamily: NMB1012-like11 d1x1ia1
17.5
6
Fold: alpha/alpha toroidSuperfamily: Chondroitin AC/alginate lyaseFamily: Hyaluronate lyase-like catalytic, N-terminal domain12 c1ky6A_
15.1
29
PDB header: endocytosis/exocytosisChain: A: PDB Molecule: alpha-adaptin c;PDBTitle: ap-2 clathrin adaptor alpha-appendage in complex with epsin2 dpw peptide
13 c3zriA_
13.2
13
PDB header: chaperoneChain: A: PDB Molecule: clpb protein;PDBTitle: n-domain of clpv from vibrio cholerae
14 d2j8wa1
12.5
17
Fold: Four-helical up-and-down bundleSuperfamily: CytochromesFamily: Cytochrome c'-like15 d1gqaa_
12.4
23
Fold: Four-helical up-and-down bundleSuperfamily: CytochromesFamily: Cytochrome c'-like16 c2c2xB_
11.5
11
PDB header: oxidoreductaseChain: B: PDB Molecule: methylenetetrahydrofolate dehydrogenase-PDBTitle: three dimensional structure of bifunctional2 methylenetetrahydrofolate dehydrogenase-cyclohydrolase3 from mycobacterium tuberculosis
17 c2ckcA_
10.7
14
PDB header: hydrolaseChain: A: PDB Molecule: chromodomain-helicase-dna-binding protein 7;PDBTitle: solution structures of the brk domains of the human chromo2 helicase domain 7 and 8, reveals structural similarity3 with gyf domain suggesting a role in protein interaction
18 d2ckca1
10.7
14
Fold: GYF/BRK domain-likeSuperfamily: BRK domain-likeFamily: BRK domain-like19 c1q2iA_
10.1
20
PDB header: antitumor proteinChain: A: PDB Molecule: pnc27;PDBTitle: nmr solution structure of a peptide from the mdm-2 binding2 domain of the p53 protein that is selectively cytotoxic to3 cancer cells
20 d1t57a_
9.0
29
Fold: Pyruvate kinase C-terminal domain-likeSuperfamily: PK C-terminal domain-likeFamily: MTH1675-like21 d1s0pa_
not modelled
8.9
18
Fold: N-terminal domain of adenylylcyclase associated protein, CAPSuperfamily: N-terminal domain of adenylylcyclase associated protein, CAPFamily: N-terminal domain of adenylylcyclase associated protein, CAP22 d1b6aa1
not modelled
8.6
19
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Methionine aminopeptidase, insert domain23 d1s05a_
not modelled
7.9
4
Fold: Four-helical up-and-down bundleSuperfamily: CytochromesFamily: Cytochrome c'-like24 d3e2ba1
not modelled
7.2
14
Fold: DLCSuperfamily: DLCFamily: DLC25 d2ikba1
not modelled
6.9
10
Fold: Lysozyme-likeSuperfamily: Lysozyme-likeFamily: NMB1012-like26 d1hdma2
not modelled
6.9
22
Fold: MHC antigen-recognition domainSuperfamily: MHC antigen-recognition domainFamily: MHC antigen-recognition domain27 c3ajfA_
not modelled
6.4
22
PDB header: viral proteinChain: A: PDB Molecule: non-structural protein 3;PDBTitle: structural insigths into dsrna binding and rna silencing suppression2 by ns3 protein of rice hoja blanca tenuivirus
28 d1pwka_
not modelled
6.3
11
Fold: DLCSuperfamily: DLCFamily: DLC29 d1k8ia2
not modelled
6.2
22
Fold: MHC antigen-recognition domainSuperfamily: MHC antigen-recognition domainFamily: MHC antigen-recognition domain30 c2o35A_
not modelled
6.2
38
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein duf1244;PDBTitle: protein of unknown function (duf1244) from sinorhizobium meliloti
31 d2o35a1
not modelled
6.2
38
Fold: SMc04008-likeSuperfamily: SMc04008-likeFamily: SMc04008-like32 d1u69a_
not modelled
6.1
44
Fold: Glyoxalase/Bleomycin resistance protein/Dihydroxybiphenyl dioxygenaseSuperfamily: Glyoxalase/Bleomycin resistance protein/Dihydroxybiphenyl dioxygenaseFamily: 3-demethylubiquinone-9 3-methyltransferase33 d1vkma_
not modelled
6.1
25
Fold: Indigoidine synthase A-likeSuperfamily: Indigoidine synthase A-likeFamily: Indigoidine synthase A-like34 d2f53d2
not modelled
6.0
24
Fold: Immunoglobulin-like beta-sandwichSuperfamily: ImmunoglobulinFamily: C1 set domains (antibody constant domain-like)35 d1cmia_
not modelled
5.8
11
Fold: DLCSuperfamily: DLCFamily: DLC36 d1vp8a_
not modelled
5.8
21
Fold: Pyruvate kinase C-terminal domain-likeSuperfamily: PK C-terminal domain-likeFamily: MTH1675-like37 d2bnqd2
not modelled
5.8
24
Fold: Immunoglobulin-like beta-sandwichSuperfamily: ImmunoglobulinFamily: C1 set domains (antibody constant domain-like)38 d1cpqa_
not modelled
5.7
13
Fold: Four-helical up-and-down bundleSuperfamily: CytochromesFamily: Cytochrome c'-like39 c3fybA_
not modelled
5.5
23
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: protein of unknown function (duf1244);PDBTitle: crystal structure of a protein of unknown function (duf1244) from2 alcanivorax borkumensis
40 d2foka4
not modelled
5.3
17
Fold: Restriction endonuclease-likeSuperfamily: Restriction endonuclease-likeFamily: Restriction endonuclease FokI, C-terminal (catalytic) domain41 c2vuxB_
not modelled
5.2
14
PDB header: oxidoreductaseChain: B: PDB Molecule: ribonucleoside-diphosphate reductase subunit m2 b;PDBTitle: human ribonucleotide reductase, subunit m2 b