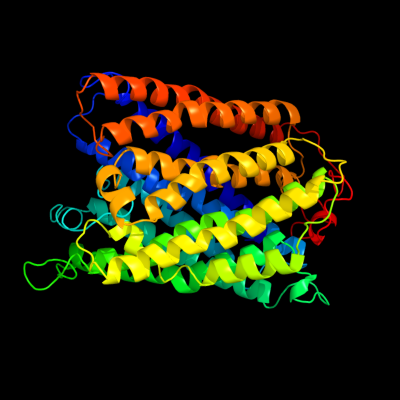
1 c1m56G_
100.0
38
PDB header: oxidoreductaseChain: G: PDB Molecule: cytochrome c oxidase;PDBTitle: structure of cytochrome c oxidase from rhodobactor2 sphaeroides (wild type)
2 d3dtua1
100.0
38
Fold: Cytochrome c oxidase subunit I-likeSuperfamily: Cytochrome c oxidase subunit I-likeFamily: Cytochrome c oxidase subunit I-like3 d1v54a_
100.0
39
Fold: Cytochrome c oxidase subunit I-likeSuperfamily: Cytochrome c oxidase subunit I-likeFamily: Cytochrome c oxidase subunit I-like4 d1ar1a_
100.0
39
Fold: Cytochrome c oxidase subunit I-likeSuperfamily: Cytochrome c oxidase subunit I-likeFamily: Cytochrome c oxidase subunit I-like5 d1ffta_
100.0
100
Fold: Cytochrome c oxidase subunit I-likeSuperfamily: Cytochrome c oxidase subunit I-likeFamily: Cytochrome c oxidase subunit I-like6 c1fftF_
100.0
100
PDB header: oxidoreductaseChain: F: PDB Molecule: ubiquinol oxidase;PDBTitle: the structure of ubiquinol oxidase from escherichia coli
7 c3eh4A_
100.0
19
PDB header: oxidoreductaseChain: A: PDB Molecule: cytochrome c oxidase subunit 1;PDBTitle: structure of the reduced form of cytochrome ba3 oxidase from thermus2 thermophilus
8 d1xmea1
100.0
20
Fold: Cytochrome c oxidase subunit I-likeSuperfamily: Cytochrome c oxidase subunit I-likeFamily: Cytochrome c oxidase subunit I-like9 c3mk7K_
100.0
14
PDB header: oxidoreductaseChain: K: PDB Molecule: cytochrome c oxidase, cbb3-type, subunit n;PDBTitle: the structure of cbb3 cytochrome oxidase
10 c3o0rB_
100.0
15
PDB header: immune system/oxidoreductaseChain: B: PDB Molecule: nitric oxide reductase subunit b;PDBTitle: crystal structure of nitric oxide reductase from pseudomonas2 aeruginosa in complex with antibody fragment
11 d1m56c_
95.2
11
Fold: Cytochrome c oxidase subunit III-likeSuperfamily: Cytochrome c oxidase subunit III-likeFamily: Cytochrome c oxidase subunit III-like12 d1v54c_
94.3
10
Fold: Cytochrome c oxidase subunit III-likeSuperfamily: Cytochrome c oxidase subunit III-likeFamily: Cytochrome c oxidase subunit III-like13 d1qlec_
93.8
10
Fold: Cytochrome c oxidase subunit III-likeSuperfamily: Cytochrome c oxidase subunit III-likeFamily: Cytochrome c oxidase subunit III-like14 d1fftb2
33.9
16
Fold: Transmembrane helix hairpinSuperfamily: Cytochrome c oxidase subunit II-like, transmembrane regionFamily: Cytochrome c oxidase subunit II-like, transmembrane region15 c3mk7F_
17.8
14
PDB header: oxidoreductaseChain: F: PDB Molecule: cytochrome c oxidase, cbb3-type, subunit p;PDBTitle: the structure of cbb3 cytochrome oxidase
16 c3chxG_
16.4
23
PDB header: membrane proteinChain: G: PDB Molecule: pmoc;PDBTitle: crystal structure of methylosinus trichosporium ob3b2 particulate methane monooxygenase (pmmo)
17 c1jauA_
16.4
38
PDB header: viral proteinChain: A: PDB Molecule: transmembrane glycoprotein (gp41);PDBTitle: nmr solution structure of the trp-rich peptide of hiv gp412 bound to dpc micelles
18 c2kncA_
13.5
11
PDB header: cell adhesionChain: A: PDB Molecule: integrin alpha-iib;PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
19 d2hf5a1
12.3
30
Fold: EF Hand-likeSuperfamily: EF-handFamily: Calmodulin-like20 d1l9bh2
11.9
43
Fold: Single transmembrane helixSuperfamily: Photosystem II reaction centre subunit H, transmembrane regionFamily: Photosystem II reaction centre subunit H, transmembrane region21 d3pvia_
not modelled
11.8
47
Fold: Restriction endonuclease-likeSuperfamily: Restriction endonuclease-likeFamily: Restriction endonuclease PvuII22 c3ke2A_
not modelled
11.6
50
PDB header: unknown functionChain: A: PDB Molecule: uncharacterized protein yp_928783.1;PDBTitle: crystal structure of a duf2131 family protein (sama_2911) from2 shewanella amazonensis sb2b at 2.50 a resolution
23 d1rzhh2
not modelled
11.4
44
Fold: Single transmembrane helixSuperfamily: Photosystem II reaction centre subunit H, transmembrane regionFamily: Photosystem II reaction centre subunit H, transmembrane region24 d2ghvc1
not modelled
11.3
20
Fold: SARS receptor-binding domain-likeSuperfamily: SARS receptor-binding domain-likeFamily: SARS receptor-binding domain-like25 d1n7za_
not modelled
10.7
23
Fold: Baseplate structural protein gp8Superfamily: Baseplate structural protein gp8Family: Baseplate structural protein gp826 d2rcrh2
not modelled
9.4
43
Fold: Single transmembrane helixSuperfamily: Photosystem II reaction centre subunit H, transmembrane regionFamily: Photosystem II reaction centre subunit H, transmembrane region27 c1yewC_
not modelled
8.5
13
PDB header: oxidoreductase, membrane proteinChain: C: PDB Molecule: particulate methane monooxygenase subunit c2;PDBTitle: crystal structure of particulate methane monooxygenase
28 c3g9rF_
not modelled
8.3
38
PDB header: viral proteinChain: F: PDB Molecule: fusion complex of hiv-1 envelope glycoproteinPDBTitle: structure of the hiv-1 gp41 membrane-proximal ectodomain2 region in a putative prefusion conformation
29 d1y5ic1
not modelled
8.3
20
Fold: Heme-binding four-helical bundleSuperfamily: Respiratory nitrate reductase 1 gamma chainFamily: Respiratory nitrate reductase 1 gamma chain30 c1javA_
not modelled
8.2
38
PDB header: viral proteinChain: A: PDB Molecule: transmembrane glycoprotein (gp41);PDBTitle: average nmr solution structure of the trp-rich peptide of2 hiv gp41 bound to dpc micelles
31 c2pv6A_
not modelled
8.0
38
PDB header: viral proteinChain: A: PDB Molecule: envelope glycoprotein;PDBTitle: hiv-1 gp41 membrane proximal ectodomain region peptide in2 dpc micelle
32 c3ir3B_
not modelled
7.3
28
PDB header: lyaseChain: B: PDB Molecule: 3-hydroxyacyl-thioester dehydratase 2;PDBTitle: crystal structure of human 3-hydroxyacyl-thioester dehydratase 22 (htd2)
33 d2qf3a1
not modelled
6.7
27
Fold: Trypsin-like serine proteasesSuperfamily: Trypsin-like serine proteasesFamily: Prokaryotic proteases34 c3kskA_
not modelled
6.7
47
PDB header: hydrolaseChain: A: PDB Molecule: type-2 restriction enzyme pvuii;PDBTitle: crystal structure of single chain pvuii
35 c2l2lB_
not modelled
6.5
26
PDB header: transferaseChain: B: PDB Molecule: methyl-cpg-binding domain protein 2;PDBTitle: solution structure of the coiled-coil complex between mbd2 and2 p66alpha
36 c1odqA_
not modelled
6.5
9
PDB header: lipid transportChain: A: PDB Molecule: apoa-i peptide;PDBTitle: peptide of human apoa-i residues 166-185. nmr, 5 structures2 at ph 3.7, 37 degrees celsius and peptide:sds mole ratio3 of 1:40
37 c1odpA_
not modelled
6.5
9
PDB header: lipid transportChain: A: PDB Molecule: apoa-i peptide;PDBTitle: peptide of human apoa-i residues 166-185. nmr, 5 structures2 at ph 6.6, 37 degrees celsius and peptide:sds mole ratio3 of 1:40
38 c1odrA_
not modelled
6.5
9
PDB header: lipid transportChain: A: PDB Molecule: apoa-i peptide;PDBTitle: peptide of human apoa-i residues 166-185. nmr, 5 structures2 at ph 6.0, 37 degrees celsius and peptide:dpc mole ratio3 of 1:40
39 c2yupA_
not modelled
6.4
18
PDB header: cell adhesionChain: A: PDB Molecule: vinexin;PDBTitle: solution structure of the second sh3 domain of human vinexin
40 d1dzfa1
not modelled
6.3
24
Fold: Restriction endonuclease-likeSuperfamily: Eukaryotic RPB5 N-terminal domainFamily: Eukaryotic RPB5 N-terminal domain41 c2fu4B_
not modelled
5.9
19
PDB header: dna binding proteinChain: B: PDB Molecule: ferric uptake regulation protein;PDBTitle: crystal structure of the dna binding domain of e.coli fur (ferric2 uptake regulator)
42 c2kncB_
not modelled
5.9
24
PDB header: cell adhesionChain: B: PDB Molecule: integrin beta-3;PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
43 d1ihwa_
not modelled
5.9
30
Fold: SH3-like barrelSuperfamily: DNA-binding domain of retroviral integraseFamily: DNA-binding domain of retroviral integrase44 d1okkd1
not modelled
5.6
21
Fold: Four-helical up-and-down bundleSuperfamily: Domain of the SRP/SRP receptor G-proteinsFamily: Domain of the SRP/SRP receptor G-proteins45 d2e74g1
not modelled
5.5
33
Fold: Single transmembrane helixSuperfamily: PetG subunit of the cytochrome b6f complexFamily: PetG subunit of the cytochrome b6f complex46 c2yu6A_
not modelled
5.5
24
PDB header: rna binding proteinChain: A: PDB Molecule: yth domain-containing protein 2;PDBTitle: solution structure of the yth domain in yth domain-2 containing protein 2
47 c1izlJ_
not modelled
5.5
21
PDB header: photosynthesisChain: J: PDB Molecule: photosystem ii: subunit psba;PDBTitle: crystal structure of photosystem ii
48 d1ctda_
not modelled
5.5
21
Fold: EF Hand-likeSuperfamily: EF-handFamily: Calmodulin-like49 d1vf5g_
not modelled
5.5
33
Fold: Single transmembrane helixSuperfamily: PetG subunit of the cytochrome b6f complexFamily: PetG subunit of the cytochrome b6f complex50 c1vf5G_
not modelled
5.5
33
PDB header: photosynthesisChain: G: PDB Molecule: protein pet g;PDBTitle: crystal structure of cytochrome b6f complex from m.laminosus
51 c1yybA_
not modelled
5.5
33
PDB header: apoptosisChain: A: PDB Molecule: programmed cell death protein 5;PDBTitle: solution structure of 1-26 fragment of human programmed2 cell death 5 protein
52 d1ehkb2
not modelled
5.4
22
Fold: Transmembrane helix hairpinSuperfamily: Cytochrome c oxidase subunit II-like, transmembrane regionFamily: Cytochrome c oxidase subunit II-like, transmembrane region53 d2j7pe1
not modelled
5.3
21
Fold: Four-helical up-and-down bundleSuperfamily: Domain of the SRP/SRP receptor G-proteinsFamily: Domain of the SRP/SRP receptor G-proteins54 d2jioa1
not modelled
5.2
25
Fold: Double psi beta-barrelSuperfamily: ADC-likeFamily: Formate dehydrogenase/DMSO reductase, C-terminal domain55 c3a1gD_
not modelled
5.0
42
PDB header: transferaseChain: D: PDB Molecule: polymerase basic protein 2;PDBTitle: high-resolution crystal structure of rna polymerase pb1-pb22 subunits from influenza a virus