| 1 |
|
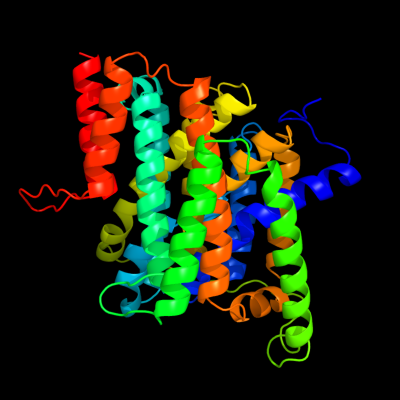
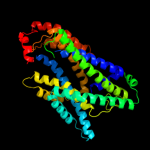
PDB 3qe7 chain A
Region: 4 - 408
Aligned: 405
Modelled: 405
Confidence: 100.0%
Identity: 100%
PDB header:transport protein
Chain: A: PDB Molecule:uracil permease;
PDBTitle: crystal structure of uracil transporter--uraa
Phyre2
| 2 |
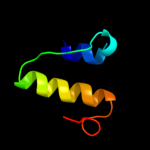
|
PDB 1dxz chain A
Region: 10 - 33
Aligned: 24
Modelled: 24
Confidence: 27.8%
Identity: 21%
PDB header:transmembrane protein
Chain: A: PDB Molecule:acetylcholine receptor protein, alpha chain;
PDBTitle: m2 transmembrane segment of alpha-subunit of nicotinic2 acetylcholine receptor from torpedo californica, nmr, 203 structures
Phyre2
| 3 |
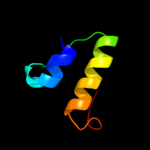
|
PDB 3lpz chain A
Region: 271 - 287
Aligned: 17
Modelled: 17
Confidence: 19.6%
Identity: 18%
PDB header:protein transport
Chain: A: PDB Molecule:get4 (yor164c homolog);
PDBTitle: crystal structure of c. therm. get4
Phyre2
| 4 |
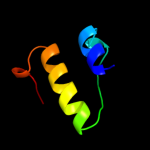
|
PDB 1v53 chain A domain 1
Region: 247 - 288
Aligned: 42
Modelled: 42
Confidence: 9.7%
Identity: 19%
Fold: Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily: Isocitrate/Isopropylmalate dehydrogenase-like
Family: Dimeric isocitrate & isopropylmalate dehydrogenases
Phyre2
| 5 |
|
PDB 1x0l chain B
Region: 247 - 288
Aligned: 42
Modelled: 42
Confidence: 8.9%
Identity: 14%
PDB header:oxidoreductase
Chain: B: PDB Molecule:homoisocitrate dehydrogenase;
PDBTitle: crystal structure of tetrameric homoisocitrate dehydrogenase from an2 extreme thermophile, thermus thermophilus
Phyre2
| 6 |
|
PDB 1g2u chain A
Region: 247 - 288
Aligned: 42
Modelled: 42
Confidence: 8.7%
Identity: 19%
Fold: Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily: Isocitrate/Isopropylmalate dehydrogenase-like
Family: Dimeric isocitrate & isopropylmalate dehydrogenases
Phyre2
| 7 |
|
PDB 1xac chain A
Region: 247 - 288
Aligned: 42
Modelled: 42
Confidence: 7.9%
Identity: 19%
Fold: Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily: Isocitrate/Isopropylmalate dehydrogenase-like
Family: Dimeric isocitrate & isopropylmalate dehydrogenases
Phyre2
| 8 |
|
PDB 3u1h chain A
Region: 247 - 288
Aligned: 42
Modelled: 42
Confidence: 7.7%
Identity: 19%
PDB header:oxidoreductase
Chain: A: PDB Molecule:3-isopropylmalate dehydrogenase;
PDBTitle: crystal structure of ipmdh from the last common ancestor of bacillus
Phyre2
| 9 |
|
PDB 3fmx chain X
Region: 247 - 288
Aligned: 42
Modelled: 42
Confidence: 7.5%
Identity: 17%
PDB header:oxidoreductase
Chain: X: PDB Molecule:tartrate dehydrogenase/decarboxylase;
PDBTitle: crystal structure of tartrate dehydrogenase from pseudomonas2 putida complexed with nadh
Phyre2
| 10 |
|
PDB 1cnz chain A
Region: 247 - 288
Aligned: 42
Modelled: 42
Confidence: 7.5%
Identity: 21%
Fold: Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily: Isocitrate/Isopropylmalate dehydrogenase-like
Family: Dimeric isocitrate & isopropylmalate dehydrogenases
Phyre2
| 11 |
|
PDB 3rj1 chain R
Region: 405 - 429
Aligned: 25
Modelled: 25
Confidence: 6.9%
Identity: 8%
PDB header:transcription
Chain: R: PDB Molecule:mediator of rna polymerase ii transcription subunit 22;
PDBTitle: architecture of the mediator head module
Phyre2
| 12 |
|
PDB 1vlc chain A
Region: 247 - 288
Aligned: 42
Modelled: 42
Confidence: 6.8%
Identity: 21%
Fold: Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily: Isocitrate/Isopropylmalate dehydrogenase-like
Family: Dimeric isocitrate & isopropylmalate dehydrogenases
Phyre2
| 13 |
|
PDB 2d4v chain D
Region: 247 - 288
Aligned: 42
Modelled: 42
Confidence: 6.7%
Identity: 7%
PDB header:oxidoreductase
Chain: D: PDB Molecule:isocitrate dehydrogenase;
PDBTitle: crystal structure of nad dependent isocitrate dehydrogenase2 from acidithiobacillus thiooxidans
Phyre2
| 14 |
|
PDB 2jln chain A
Region: 72 - 426
Aligned: 353
Modelled: 355
Confidence: 6.5%
Identity: 11%
PDB header:membrane protein
Chain: A: PDB Molecule:mhp1;
PDBTitle: structure of mhp1, a nucleobase-cation-symport-1 family2 transporter
Phyre2
| 15 |
|
PDB 1cm7 chain A
Region: 247 - 288
Aligned: 42
Modelled: 42
Confidence: 6.5%
Identity: 21%
Fold: Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily: Isocitrate/Isopropylmalate dehydrogenase-like
Family: Dimeric isocitrate & isopropylmalate dehydrogenases
Phyre2
| 16 |
|
PDB 1wpw chain A
Region: 247 - 288
Aligned: 42
Modelled: 42
Confidence: 6.5%
Identity: 21%
Fold: Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily: Isocitrate/Isopropylmalate dehydrogenase-like
Family: Dimeric isocitrate & isopropylmalate dehydrogenases
Phyre2
| 17 |
|
PDB 1w0d chain A
Region: 247 - 288
Aligned: 42
Modelled: 42
Confidence: 6.5%
Identity: 10%
Fold: Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily: Isocitrate/Isopropylmalate dehydrogenase-like
Family: Dimeric isocitrate & isopropylmalate dehydrogenases
Phyre2
| 18 |
|
PDB 1hqs chain A
Region: 247 - 288
Aligned: 42
Modelled: 42
Confidence: 6.4%
Identity: 10%
Fold: Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily: Isocitrate/Isopropylmalate dehydrogenase-like
Family: Dimeric isocitrate & isopropylmalate dehydrogenases
Phyre2
| 19 |
|
PDB 1a05 chain A
Region: 247 - 288
Aligned: 42
Modelled: 42
Confidence: 6.0%
Identity: 21%
Fold: Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily: Isocitrate/Isopropylmalate dehydrogenase-like
Family: Dimeric isocitrate & isopropylmalate dehydrogenases
Phyre2
| 20 |
|
PDB 2d1c chain B
Region: 247 - 288
Aligned: 42
Modelled: 42
Confidence: 5.9%
Identity: 19%
PDB header:oxidoreductase
Chain: B: PDB Molecule:isocitrate dehydrogenase;
PDBTitle: crystal structure of tt0538 protein from thermus thermophilus hb8
Phyre2
| 21 |
|
| 22 |
|