| 1 |
|
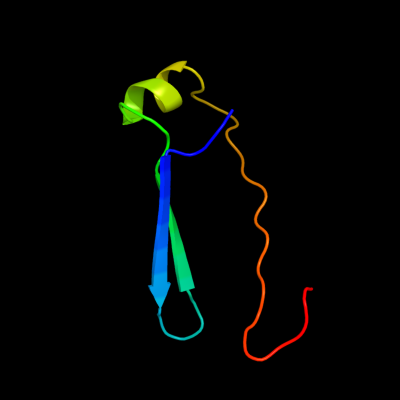
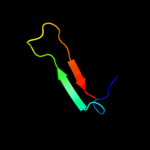
PDB 1y6k chain R domain 2
Region: 50 - 99
Aligned: 50
Modelled: 50
Confidence: 23.4%
Identity: 24%
Fold: Immunoglobulin-like beta-sandwich
Superfamily: Fibronectin type III
Family: Fibronectin type III
Phyre2
| 2 |
|
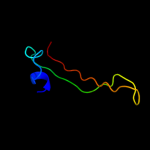
PDB 3gqq chain D
Region: 57 - 74
Aligned: 18
Modelled: 15
Confidence: 19.4%
Identity: 50%
PDB header:splicing
Chain: D: PDB Molecule:protein unc-119 homolog a;
PDBTitle: crystal structure of the human retinal protein 4 (unc-1192 homolog a). northeast structural genomics consortium3 target hr3066a
Phyre2
| 3 |
|
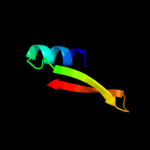
PDB 1sdw chain A domain 2
Region: 45 - 73
Aligned: 29
Modelled: 29
Confidence: 18.3%
Identity: 17%
Fold: Nucleoplasmin-like/VP (viral coat and capsid proteins)
Superfamily: PHM/PNGase F
Family: Peptidylglycine alpha-hydroxylating monooxygenase, PHM
Phyre2
| 4 |
|
PDB 2k8j chain X
Region: 30 - 38
Aligned: 9
Modelled: 9
Confidence: 13.7%
Identity: 78%
PDB header:viral protein
Chain: X: PDB Molecule:p7tm2;
PDBTitle: solution structure of hcv p7 tm2
Phyre2
| 5 |
|
PDB 2k2w chain A
Region: 30 - 44
Aligned: 15
Modelled: 15
Confidence: 12.4%
Identity: 27%
PDB header:cell cycle
Chain: A: PDB Molecule:recombination and dna repair protein;
PDBTitle: second brct domain of nbs1
Phyre2
| 6 |
|
PDB 1r4x chain A domain 1
Region: 35 - 75
Aligned: 41
Modelled: 41
Confidence: 9.3%
Identity: 22%
Fold: Immunoglobulin-like beta-sandwich
Superfamily: Clathrin adaptor appendage domain
Family: Coatomer appendage domain
Phyre2
| 7 |
|
PDB 2lka chain A
Region: 43 - 47
Aligned: 5
Modelled: 5
Confidence: 9.2%
Identity: 100%
PDB header:toxin
Chain: A: PDB Molecule:toxin ts16;
PDBTitle: new tricks of an old fold: structural versatility of scorpion toxins2 with common cysteine spacing
Phyre2
| 8 |
|
PDB 2rnr chain B domain 1
Region: 2 - 27
Aligned: 26
Modelled: 26
Confidence: 9.1%
Identity: 23%
Fold: PH domain-like barrel
Superfamily: PH domain-like
Family: TFIIH domain
Phyre2
| 9 |
|
PDB 2li3 chain A
Region: 43 - 47
Aligned: 5
Modelled: 5
Confidence: 8.3%
Identity: 100%
PDB header:toxin
Chain: A: PDB Molecule:potassium channel toxin kappa-ktx3.1;
PDBTitle: structural and functional analysis of a novel potassium toxin2 argentinean scorpion tityus trivittatus reveals a new kappa sub-3 family
Phyre2
| 10 |
|
PDB 1pzd chain A domain 1
Region: 35 - 75
Aligned: 41
Modelled: 41
Confidence: 7.6%
Identity: 22%
Fold: Immunoglobulin-like beta-sandwich
Superfamily: Clathrin adaptor appendage domain
Family: Coatomer appendage domain
Phyre2
| 11 |
|
PDB 1umg chain A
Region: 31 - 53
Aligned: 23
Modelled: 23
Confidence: 6.3%
Identity: 43%
Fold: Sulfolobus fructose-1,6-bisphosphatase-like
Superfamily: Sulfolobus fructose-1,6-bisphosphatase-like
Family: Sulfolobus fructose-1,6-bisphosphatase-like
Phyre2
| 12 |
|
PDB 1opm chain A
Region: 45 - 73
Aligned: 29
Modelled: 29
Confidence: 6.2%
Identity: 17%
PDB header:oxidoreductase
Chain: A: PDB Molecule:protein (peptidylglycine alpha-hydroxylating
PDBTitle: oxidized (cu2+) peptidylglycine alpha-hydroxylating monooxygenase2 (phm) with bound substrate
Phyre2
| 13 |
|
PDB 2dvk chain A
Region: 38 - 69
Aligned: 32
Modelled: 32
Confidence: 5.7%
Identity: 31%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:upf0130 protein ape0816;
PDBTitle: crystal structure of hypothetical protein from aeropyrum pernix
Phyre2
| 14 |
|
PDB 1pzd chain A
Region: 49 - 75
Aligned: 27
Modelled: 27
Confidence: 5.7%
Identity: 30%
PDB header:endocytosis/exocytosis
Chain: A: PDB Molecule:coatomer gamma subunit;
PDBTitle: structural identification of a conserved appendage domain2 in the carboxyl-terminus of the copi gamma-subunit.
Phyre2