| 1 |
|
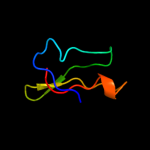
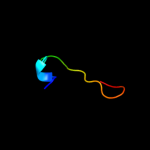
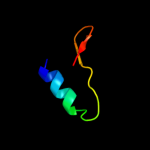
PDB 3bbo chain 6
Region: 1 - 41
Aligned: 38
Modelled: 41
Confidence: 99.1%
Identity: 55%
PDB header:ribosome
Chain: 6: PDB Molecule:ribosomal protein l36;
PDBTitle: homology model for the spinach chloroplast 50s subunit2 fitted to 9.4a cryo-em map of the 70s chlororibosome
Phyre2
| 2 |
|
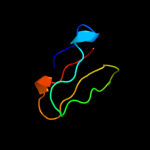
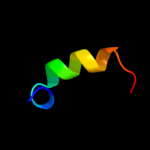
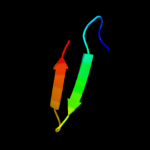
PDB 2qbk chain 4
Region: 1 - 41
Aligned: 38
Modelled: 41
Confidence: 98.8%
Identity: 47%
PDB header:ribosome
Chain: 4: PDB Molecule:50s ribosomal protein l36;
PDBTitle: crystal structure of the bacterial ribosome from escherichia2 coli in complex with gentamicin and ribosome recycling3 factor (rrf). this file contains the 50s subunit of the4 second 70s ribosome, with gentamicin and rrf bound. the5 entire crystal structure contains two 70s ribosomes and is6 described in remark 400.
Phyre2
| 3 |
|
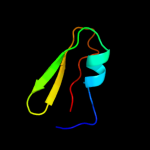
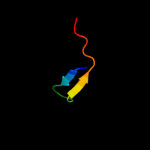
PDB 1dfe chain A
Region: 1 - 41
Aligned: 37
Modelled: 41
Confidence: 98.7%
Identity: 49%
Fold: Ribosomal protein L36
Superfamily: Ribosomal protein L36
Family: Ribosomal protein L36
Phyre2
| 4 |
|
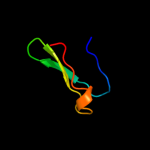
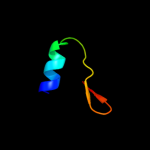
PDB 2zjr chain 4 domain 1
Region: 9 - 41
Aligned: 32
Modelled: 33
Confidence: 98.4%
Identity: 44%
Fold: Ribosomal protein L36
Superfamily: Ribosomal protein L36
Family: Ribosomal protein L36
Phyre2
| 5 |
|
PDB 1w7a chain B domain 4
Region: 9 - 27
Aligned: 19
Modelled: 19
Confidence: 16.2%
Identity: 32%
Fold: MutS N-terminal domain-like
Superfamily: DNA repair protein MutS, domain I
Family: DNA repair protein MutS, domain I
Phyre2
| 6 |
|
PDB 1ni8 chain A
Region: 2 - 19
Aligned: 18
Modelled: 18
Confidence: 13.3%
Identity: 33%
Fold: H-NS histone-like proteins
Superfamily: H-NS histone-like proteins
Family: H-NS histone-like proteins
Phyre2
| 7 |
|
PDB 1cz6 chain A
Region: 15 - 34
Aligned: 20
Modelled: 20
Confidence: 8.2%
Identity: 30%
PDB header:toxin
Chain: A: PDB Molecule:protein (androctonin);
PDBTitle: solution structure of androctonin
Phyre2
| 8 |
|
PDB 1wb9 chain A domain 4
Region: 4 - 29
Aligned: 26
Modelled: 26
Confidence: 6.0%
Identity: 23%
Fold: MutS N-terminal domain-like
Superfamily: DNA repair protein MutS, domain I
Family: DNA repair protein MutS, domain I
Phyre2
| 9 |
|
PDB 1l4d chain B
Region: 15 - 31
Aligned: 17
Modelled: 17
Confidence: 5.9%
Identity: 35%
Fold: beta-Grasp (ubiquitin-like)
Superfamily: Staphylokinase/streptokinase
Family: Staphylokinase/streptokinase
Phyre2
| 10 |
|
PDB 1ewq chain A domain 4
Region: 3 - 29
Aligned: 27
Modelled: 27
Confidence: 5.9%
Identity: 19%
Fold: MutS N-terminal domain-like
Superfamily: DNA repair protein MutS, domain I
Family: DNA repair protein MutS, domain I
Phyre2
| 11 |
|
PDB 1ni5 chain A domain 4
Region: 13 - 30
Aligned: 18
Modelled: 18
Confidence: 5.7%
Identity: 22%
Fold: MesJ substrate recognition domain-like
Superfamily: MesJ substrate recognition domain-like
Family: MesJ substrate recognition domain-like
Phyre2
| 12 |
|
PDB 1l4z chain B
Region: 15 - 31
Aligned: 17
Modelled: 17
Confidence: 5.5%
Identity: 35%
Fold: beta-Grasp (ubiquitin-like)
Superfamily: Staphylokinase/streptokinase
Family: Staphylokinase/streptokinase
Phyre2