| 1 |
|
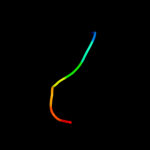
PDB 2haf chain A domain 1
Region: 26 - 33
Aligned: 8
Modelled: 8
Confidence: 20.0%
Identity: 63%
Fold: VC0467-like
Superfamily: VC0467-like
Family: VC0467-like
Phyre2
| 2 |
|
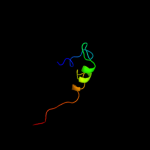
PDB 2aj2 chain A
Region: 26 - 33
Aligned: 8
Modelled: 8
Confidence: 17.7%
Identity: 63%
PDB header:unknown function
Chain: A: PDB Molecule:hypothetical upf0301 protein vc0467;
PDBTitle: x-ray crystal structure of protein vc0467 from vibrio2 cholerae. northeast structural genomics consortium target3 vcr8.
Phyre2
| 3 |
|
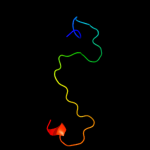
PDB 2do8 chain A domain 1
Region: 26 - 33
Aligned: 8
Modelled: 8
Confidence: 16.5%
Identity: 38%
Fold: VC0467-like
Superfamily: VC0467-like
Family: VC0467-like
Phyre2
| 4 |
|
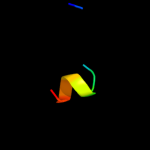
PDB 2gs5 chain A domain 1
Region: 26 - 33
Aligned: 8
Modelled: 8
Confidence: 15.7%
Identity: 25%
Fold: VC0467-like
Superfamily: VC0467-like
Family: VC0467-like
Phyre2
| 5 |
|
PDB 2ew0 chain A domain 1
Region: 26 - 33
Aligned: 8
Modelled: 8
Confidence: 13.2%
Identity: 50%
Fold: VC0467-like
Superfamily: VC0467-like
Family: VC0467-like
Phyre2
| 6 |
|
PDB 2gzo chain A domain 1
Region: 26 - 33
Aligned: 8
Modelled: 8
Confidence: 12.7%
Identity: 50%
Fold: VC0467-like
Superfamily: VC0467-like
Family: VC0467-like
Phyre2
| 7 |
|
PDB 2g7h chain A
Region: 13 - 60
Aligned: 40
Modelled: 45
Confidence: 12.3%
Identity: 33%
PDB header:transferase
Chain: A: PDB Molecule:methylated-dna--protein-cysteine
PDBTitle: structure of an o6-methylguanine dna methyltransferase from2 methanococcus jannaschii (mj1529)
Phyre2
| 8 |
|
PDB 3izb chain T
Region: 53 - 63
Aligned: 11
Modelled: 11
Confidence: 11.9%
Identity: 36%
PDB header:ribosome
Chain: T: PDB Molecule:40s ribosomal protein rps21 (s21e);
PDBTitle: localization of the small subunit ribosomal proteins into a 6.1 a2 cryo-em map of saccharomyces cerevisiae translating 80s ribosome
Phyre2
| 9 |
|
PDB 1jdq chain A
Region: 40 - 75
Aligned: 36
Modelled: 36
Confidence: 9.4%
Identity: 25%
Fold: IF3-like
Superfamily: SirA-like
Family: SirA-like
Phyre2
| 10 |
|
PDB 1kn0 chain A
Region: 26 - 32
Aligned: 7
Modelled: 7
Confidence: 5.6%
Identity: 71%
Fold: dsRBD-like
Superfamily: dsRNA-binding domain-like
Family: The homologous-pairing domain of Rad52 recombinase
Phyre2
| 11 |
|
PDB 3iz6 chain T
Region: 53 - 63
Aligned: 11
Modelled: 11
Confidence: 5.6%
Identity: 45%
PDB header:ribosome
Chain: T: PDB Molecule:40s ribosomal protein s21 (s21e);
PDBTitle: localization of the small subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
Phyre2
| 12 |
|
PDB 1yvr chain A domain 1
Region: 30 - 39
Aligned: 10
Modelled: 10
Confidence: 5.5%
Identity: 50%
Fold: alpha-alpha superhelix
Superfamily: TROVE domain-like
Family: TROVE domain-like
Phyre2