| 1 |
|
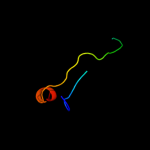
PDB 1cks chain A
Region: 116 - 124
Aligned: 9
Modelled: 9
Confidence: 35.9%
Identity: 67%
Fold: Cell cycle regulatory proteins
Superfamily: Cell cycle regulatory proteins
Family: Cell cycle regulatory proteins
Phyre2
| 2 |
|
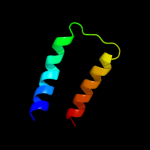
PDB 2ast chain C domain 1
Region: 116 - 124
Aligned: 9
Modelled: 9
Confidence: 35.1%
Identity: 67%
Fold: Cell cycle regulatory proteins
Superfamily: Cell cycle regulatory proteins
Family: Cell cycle regulatory proteins
Phyre2
| 3 |
|
PDB 1puc chain A
Region: 116 - 124
Aligned: 9
Modelled: 9
Confidence: 33.3%
Identity: 56%
Fold: Cell cycle regulatory proteins
Superfamily: Cell cycle regulatory proteins
Family: Cell cycle regulatory proteins
Phyre2
| 4 |
|
PDB 1qb3 chain B
Region: 116 - 124
Aligned: 9
Modelled: 9
Confidence: 31.5%
Identity: 56%
PDB header:cell cycle
Chain: B: PDB Molecule:cyclin-dependent kinases regulatory subunit;
PDBTitle: crystal structure of the cell cycle regulatory protein cks1
Phyre2
| 5 |
|
PDB 1qb3 chain A
Region: 116 - 124
Aligned: 9
Modelled: 9
Confidence: 30.6%
Identity: 56%
Fold: Cell cycle regulatory proteins
Superfamily: Cell cycle regulatory proteins
Family: Cell cycle regulatory proteins
Phyre2
| 6 |
|
PDB 2nvn chain A domain 1
Region: 117 - 134
Aligned: 18
Modelled: 18
Confidence: 10.4%
Identity: 28%
Fold: ssDNA-binding transcriptional regulator domain
Superfamily: ssDNA-binding transcriptional regulator domain
Family: PMN2A0962/syc2379c-like
Phyre2
| 7 |
|
PDB 2it9 chain A domain 1
Region: 117 - 134
Aligned: 18
Modelled: 18
Confidence: 9.7%
Identity: 28%
Fold: ssDNA-binding transcriptional regulator domain
Superfamily: ssDNA-binding transcriptional regulator domain
Family: PMN2A0962/syc2379c-like
Phyre2
| 8 |
|
PDB 1wq6 chain A
Region: 118 - 128
Aligned: 11
Modelled: 11
Confidence: 8.0%
Identity: 36%
PDB header:oncoprotein
Chain: A: PDB Molecule:aml1-eto;
PDBTitle: the tetramer structure of the nervy homolgy two (nhr2) domain of aml1-2 eto is critical for aml1-eto's activity
Phyre2
| 9 |
|
PDB 1na6 chain A domain 1
Region: 92 - 120
Aligned: 29
Modelled: 29
Confidence: 7.7%
Identity: 28%
Fold: DNA-binding pseudobarrel domain
Superfamily: DNA-binding pseudobarrel domain
Family: Type II restriction endonuclease effector domain
Phyre2
| 10 |
|
PDB 3cx5 chain C domain 1
Region: 15 - 57
Aligned: 43
Modelled: 43
Confidence: 6.3%
Identity: 16%
Fold: a domain/subunit of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)
Superfamily: a domain/subunit of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)
Family: a domain/subunit of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)
Phyre2
| 11 |
|
PDB 1d1d chain A domain 1
Region: 49 - 59
Aligned: 11
Modelled: 11
Confidence: 5.3%
Identity: 36%
Fold: Acyl carrier protein-like
Superfamily: Retrovirus capsid dimerization domain-like
Family: Retrovirus capsid protein C-terminal domain
Phyre2
| 12 |
|
PDB 1in1 chain A
Region: 50 - 58
Aligned: 9
Modelled: 9
Confidence: 5.3%
Identity: 33%
Fold: BRCT domain
Superfamily: BRCT domain
Family: DNA ligase
Phyre2
| 13 |
|
PDB 1z9e chain A
Region: 52 - 66
Aligned: 15
Modelled: 15
Confidence: 5.3%
Identity: 7%
PDB header:protein binding/transcription
Chain: A: PDB Molecule:pc4 and sfrs1 interacting protein 2;
PDBTitle: solution structure of the hiv-1 integrase-binding domain in2 ledgf/p75
Phyre2
| 14 |
|
PDB 3c8i chain A
Region: 112 - 133
Aligned: 22
Modelled: 22
Confidence: 5.1%
Identity: 18%
PDB header:membrane protein
Chain: A: PDB Molecule:putative membrane protein;
PDBTitle: crystal structure of a putative membrane protein from corynebacterium2 diphtheriae
Phyre2