| 1 |
|
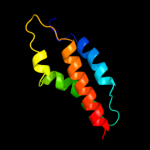
PDB 1tlq chain A
Region: 86 - 162
Aligned: 68
Modelled: 77
Confidence: 99.9%
Identity: 28%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein ypjq;
PDBTitle: crystal structure of protein ypjq from bacillus subtilis, pfam duf64
Phyre2
| 2 |
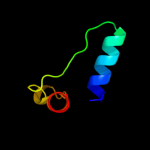
|
PDB 1tlq chain A
Region: 86 - 162
Aligned: 68
Modelled: 77
Confidence: 99.9%
Identity: 28%
Fold: YutG-like
Superfamily: YutG-like
Family: YutG-like
Phyre2
| 3 |
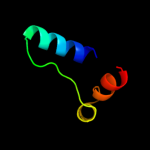
|
PDB 1rfz chain A
Region: 86 - 164
Aligned: 71
Modelled: 79
Confidence: 99.9%
Identity: 23%
Fold: YutG-like
Superfamily: YutG-like
Family: YutG-like
Phyre2
| 4 |
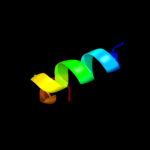
|
PDB 1y9i chain A
Region: 84 - 165
Aligned: 74
Modelled: 82
Confidence: 99.9%
Identity: 20%
Fold: YutG-like
Superfamily: YutG-like
Family: YutG-like
Phyre2
| 5 |
|
PDB 1loi chain A
Region: 127 - 136
Aligned: 10
Modelled: 10
Confidence: 15.2%
Identity: 50%
PDB header:hydrolase
Chain: A: PDB Molecule:cyclic 3',5'-amp specific phosphodiesterase rd1;
PDBTitle: n-terminal splice region of rat c-amp phosphodiesterase,2 nmr, 7 structures
Phyre2
| 6 |
|
PDB 2zxi chain C
Region: 111 - 150
Aligned: 36
Modelled: 40
Confidence: 12.9%
Identity: 28%
PDB header:fad-binding protein
Chain: C: PDB Molecule:trna uridine 5-carboxymethylaminomethyl
PDBTitle: structure of aquifex aeolicus gida in the form ii crystal
Phyre2
| 7 |
|
PDB 3cp8 chain C
Region: 111 - 150
Aligned: 36
Modelled: 40
Confidence: 10.0%
Identity: 22%
PDB header:oxidoreductase
Chain: C: PDB Molecule:trna uridine 5-carboxymethylaminomethyl
PDBTitle: crystal structure of gida from chlorobium tepidum
Phyre2
| 8 |
|
PDB 1ctd chain A
Region: 119 - 124
Aligned: 6
Modelled: 6
Confidence: 9.6%
Identity: 67%
Fold: EF Hand-like
Superfamily: EF-hand
Family: Calmodulin-like
Phyre2
| 9 |
|
PDB 2b6c chain A domain 1
Region: 74 - 91
Aligned: 18
Modelled: 18
Confidence: 7.9%
Identity: 0%
Fold: alpha-alpha superhelix
Superfamily: ARM repeat
Family: BC3264-like
Phyre2
| 10 |
|
PDB 3eeq chain B
Region: 74 - 94
Aligned: 21
Modelled: 21
Confidence: 7.4%
Identity: 10%
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:putative cobalamin biosynthesis protein g
PDBTitle: crystal structure of a putative cobalamin biosynthesis2 protein g homolog from sulfolobus solfataricus
Phyre2
| 11 |
|
PDB 3eeq chain A domain 2
Region: 83 - 94
Aligned: 12
Modelled: 12
Confidence: 5.8%
Identity: 17%
Fold: CbiG N-terminal domain-like
Superfamily: CbiG N-terminal domain-like
Family: CbiG N-terminal domain-like
Phyre2