| 1 |
|
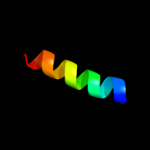
PDB 2g3v chain C
Region: 43 - 100
Aligned: 58
Modelled: 56
Confidence: 16.7%
Identity: 21%
PDB header:unknown function
Chain: C: PDB Molecule:cag pathogenicity island protein 13;
PDBTitle: crystal structure of cags (hp0534, cag13) from helicobacter2 pylori
Phyre2
| 2 |
|
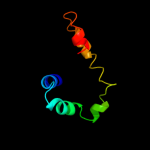
PDB 2zsh chain B
Region: 116 - 127
Aligned: 12
Modelled: 12
Confidence: 11.9%
Identity: 50%
PDB header:hormone receptor
Chain: B: PDB Molecule:della protein gai;
PDBTitle: structural basis of gibberellin(ga3)-induced della2 recognition by the gibberellin receptor
Phyre2
| 3 |
|
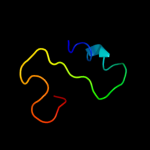
PDB 1wpg chain A domain 4
Region: 12 - 62
Aligned: 48
Modelled: 51
Confidence: 11.8%
Identity: 23%
Fold: Calcium ATPase, transmembrane domain M
Superfamily: Calcium ATPase, transmembrane domain M
Family: Calcium ATPase, transmembrane domain M
Phyre2
| 4 |
|
PDB 2inc chain B domain 1
Region: 82 - 109
Aligned: 27
Modelled: 28
Confidence: 10.5%
Identity: 22%
Fold: Ferritin-like
Superfamily: Ferritin-like
Family: Ribonucleotide reductase-like
Phyre2
| 5 |
|
PDB 1kf6 chain D
Region: 12 - 80
Aligned: 60
Modelled: 64
Confidence: 10.1%
Identity: 35%
Fold: Heme-binding four-helical bundle
Superfamily: Fumarate reductase respiratory complex transmembrane subunits
Family: Succinate dehydrogenase/Fumarate reductase transmembrane subunits (SdhC/FrdC and SdhD/FrdD)
Phyre2
| 6 |
|
PDB 3e9v chain A domain 1
Region: 39 - 68
Aligned: 30
Modelled: 30
Confidence: 8.0%
Identity: 13%
Fold: BTG domain-like
Superfamily: BTG domain-like
Family: BTG domain-like
Phyre2
| 7 |
|
PDB 1p58 chain C
Region: 120 - 156
Aligned: 30
Modelled: 37
Confidence: 7.4%
Identity: 37%
PDB header:virus
Chain: C: PDB Molecule:major envelope protein e;
PDBTitle: complex organization of dengue virus membrane proteins as revealed by2 9.5 angstrom cryo-em reconstruction
Phyre2
| 8 |
|
PDB 1w2z chain A domain 3
Region: 104 - 118
Aligned: 15
Modelled: 15
Confidence: 6.4%
Identity: 7%
Fold: Cystatin-like
Superfamily: Amine oxidase N-terminal region
Family: Amine oxidase N-terminal region
Phyre2
| 9 |
|
PDB 1w6g chain A domain 3
Region: 104 - 118
Aligned: 15
Modelled: 15
Confidence: 5.9%
Identity: 27%
Fold: Cystatin-like
Superfamily: Amine oxidase N-terminal region
Family: Amine oxidase N-terminal region
Phyre2
| 10 |
|
PDB 2o01 chain D
Region: 84 - 115
Aligned: 32
Modelled: 32
Confidence: 5.7%
Identity: 28%
PDB header:photosynthesis
Chain: D: PDB Molecule:photosystem i reaction center subunit ii,
PDBTitle: the structure of a plant photosystem i supercomplex at 3.42 angstrom resolution
Phyre2
| 11 |
|
PDB 2a3z chain C
Region: 171 - 178
Aligned: 8
Modelled: 8
Confidence: 5.6%
Identity: 50%
PDB header:structural protein
Chain: C: PDB Molecule:wiskott-aldrich syndrome protein;
PDBTitle: ternary complex of the wh2 domain of wasp with actin-dnase i
Phyre2
| 12 |
|
PDB 2z15 chain A domain 1
Region: 39 - 68
Aligned: 30
Modelled: 30
Confidence: 5.6%
Identity: 20%
Fold: BTG domain-like
Superfamily: BTG domain-like
Family: BTG domain-like
Phyre2
| 13 |
|
PDB 1d6z chain A domain 3
Region: 104 - 118
Aligned: 15
Modelled: 15
Confidence: 5.4%
Identity: 20%
Fold: Cystatin-like
Superfamily: Amine oxidase N-terminal region
Family: Amine oxidase N-terminal region
Phyre2
| 14 |
|
PDB 2p7t chain C domain 1
Region: 130 - 146
Aligned: 17
Modelled: 17
Confidence: 5.4%
Identity: 29%
Fold: Voltage-gated potassium channels
Superfamily: Voltage-gated potassium channels
Family: Voltage-gated potassium channels
Phyre2
| 15 |
|
PDB 1hlm chain A
Region: 35 - 102
Aligned: 66
Modelled: 68
Confidence: 5.1%
Identity: 8%
Fold: Globin-like
Superfamily: Globin-like
Family: Globins
Phyre2
| 16 |
|
PDB 2wse chain D
Region: 84 - 115
Aligned: 32
Modelled: 32
Confidence: 5.1%
Identity: 34%
PDB header:photosynthesis
Chain: D: PDB Molecule:photosystem i reaction center subunit ii,
PDBTitle: improved model of plant photosystem i
Phyre2