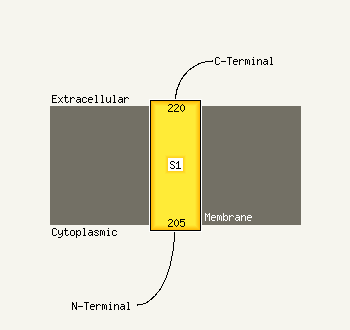
1 d1utaa_
98.0
18
Fold: Ferredoxin-likeSuperfamily: Sporulation related repeatFamily: Sporulation related repeat2 c1x60A_
97.9
20
PDB header: hydrolaseChain: A: PDB Molecule: sporulation-specific n-acetylmuramoyl-l-alaninePDBTitle: solution structure of the peptidoglycan binding domain of2 b. subtilis cell wall lytic enzyme cwlc
3 c3g5oA_
32.8
23
PDB header: toxin/antitoxinChain: A: PDB Molecule: uncharacterized protein rv2865;PDBTitle: the crystal structure of the toxin-antitoxin complex relbe2 (rv2865-2 2866) from mycobacterium tuberculosis
4 c3kgkA_
25.3
13
PDB header: chaperoneChain: A: PDB Molecule: arsenical resistance operon trans-acting repressor arsd;PDBTitle: crystal structure of arsd
5 c3ktbD_
24.3
13
PDB header: transcription regulatorChain: D: PDB Molecule: arsenical resistance operon trans-acting repressor;PDBTitle: crystal structure of arsenical resistance operon trans-acting2 repressor from bacteroides vulgatus atcc 8482
6 d1xrsb2
20.0
17
Fold: Dodecin subunit-likeSuperfamily: D-lysine 5,6-aminomutase beta subunit KamE, N-terminal domainFamily: D-lysine 5,6-aminomutase beta subunit KamE, N-terminal domain7 d2k49a1
19.1
20
Fold: YegP-likeSuperfamily: YegP-likeFamily: YegP-like8 d1qqka_
15.8
14
Fold: beta-TrefoilSuperfamily: CytokineFamily: Fibroblast growth factors (FGF)9 c2odkD_
13.6
29
PDB header: structural genomics, unknown functionChain: D: PDB Molecule: hypothetical protein;PDBTitle: putative prevent-host-death protein from nitrosomonas europaea
10 c1ywlA_
12.5
14
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical upf0213 protein ef2693;PDBTitle: solution nmr structure of the protein ef2693 from e.2 faecalis: northeast structural genomics consortium target3 efr36
11 d2odka1
12.5
29
Fold: YefM-likeSuperfamily: YefM-likeFamily: YefM-like12 c2jvrA_
12.5
20
PDB header: rna binding proteinChain: A: PDB Molecule: nucleolar protein 3;PDBTitle: segmental isotope labeling of npl3p
13 d2a6qa1
12.4
21
Fold: YefM-likeSuperfamily: YefM-likeFamily: YefM-like14 d2cqia1
11.7
12
Fold: Ferredoxin-likeSuperfamily: RNA-binding domain, RBDFamily: Canonical RBD15 d1tuaa2
11.6
33
Fold: Eukaryotic type KH-domain (KH-domain type I)Superfamily: Eukaryotic type KH-domain (KH-domain type I)Family: Eukaryotic type KH-domain (KH-domain type I)16 c2osrA_
11.2
19
PDB header: rna binding proteinChain: A: PDB Molecule: nucleolar protein 3;PDBTitle: nmr structure of rrm-2 of yeast npl3 protein
17 d2k7ia1
10.5
17
Fold: YegP-likeSuperfamily: YegP-likeFamily: YegP-like18 c2k7iB_
10.5
17
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: upf0339 protein atu0232;PDBTitle: solution nmr structure of protein atu0232 from agrobacterium2 tumefaciens. northeast structural genomics consortium (nesg) target3 att3. ontario center for structural proteomics target atc0223.
19 d1x5sa1
10.1
21
Fold: Ferredoxin-likeSuperfamily: RNA-binding domain, RBDFamily: Canonical RBD20 c2dnzA_
9.7
17
PDB header: rna binding proteinChain: A: PDB Molecule: probable rna-binding protein 23;PDBTitle: solution structure of the second rna binding domain of rna2 binding motif protein 23
21 d1ihka_
not modelled
8.6
17
Fold: beta-TrefoilSuperfamily: CytokineFamily: Fibroblast growth factors (FGF)22 c3jtzA_
not modelled
8.3
19
PDB header: dna binding proteinChain: A: PDB Molecule: integrase;PDBTitle: structure of the arm-type binding domain of hpi integrase
23 c3ju0A_
not modelled
8.3
20
PDB header: dna binding proteinChain: A: PDB Molecule: phage integrase;PDBTitle: structure of the arm-type binding domain of hai7 integrase
24 c2dnmA_
not modelled
8.0
15
PDB header: rna binding proteinChain: A: PDB Molecule: srp46 splicing factor;PDBTitle: solution structure of rna binding domain in srp46 splicing2 factor
25 c2wbrA_
not modelled
7.9
24
PDB header: dna-binding proteinChain: A: PDB Molecule: gw182;PDBTitle: the rrm domain in gw182 proteins contributes to mirna-2 mediated gene silencing
26 c2dh9A_
not modelled
7.6
13
PDB header: rna binding proteinChain: A: PDB Molecule: heterogeneous nuclear ribonucleoprotein m;PDBTitle: solution structure of the c-terminal rna binding domain in2 heterogeneous nuclear ribonucleoprotein m
27 c1vjqB_
not modelled
7.3
27
PDB header: structural genomics, de novo proteinChain: B: PDB Molecule: designed protein;PDBTitle: designed protein based on backbone conformation of2 procarboxypeptidase-a (1aye) with sidechains chosen for maximal3 predicted stability.
28 c2oq8A_
not modelled
6.8
33
PDB header: dna binding proteinChain: A: PDB Molecule: hypothetical protein;PDBTitle: structure of orf 157 from acidianus filamentous virus 1
29 d3e2ba1
not modelled
6.5
18
Fold: DLCSuperfamily: DLCFamily: DLC30 c3femB_
not modelled
6.5
15
PDB header: biosynthetic protein, transferaseChain: B: PDB Molecule: pyridoxine biosynthesis protein snz1;PDBTitle: structure of the synthase subunit pdx1.1 (snz1) of plp synthase from2 saccharomyces cerevisiae
31 c2k49A_
not modelled
6.4
12
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: upf0339 protein so_3888;PDBTitle: solution nmr structure of upf0339 protein so3888 from shewanella2 oneidensis. northeast structural genomics consortium target sor190
32 d1vkwa_
not modelled
6.4
31
Fold: FMN-dependent nitroreductase-likeSuperfamily: FMN-dependent nitroreductase-likeFamily: Putative nitroreductase TM158633 c3qbtH_
not modelled
6.3
16
PDB header: protein transport/hydrolaseChain: H: PDB Molecule: inositol polyphosphate 5-phosphatase ocrl-1;PDBTitle: crystal structure of ocrl1 540-678 in complex with rab8a:gppnhp
34 d1kr4a_
not modelled
6.0
25
Fold: Ferredoxin-likeSuperfamily: GlnB-likeFamily: Divalent ion tolerance proteins CutA (CutA1)35 c2rneA_
not modelled
5.8
10
PDB header: rna binding proteinChain: A: PDB Molecule: tia1 protein;PDBTitle: solution structure of the second rna recognition motif2 (rrm) of tia-1
36 d2cq3a1
not modelled
5.7
14
Fold: Ferredoxin-likeSuperfamily: RNA-binding domain, RBDFamily: Canonical RBD37 d1nuna_
not modelled
5.6
24
Fold: beta-TrefoilSuperfamily: CytokineFamily: Fibroblast growth factors (FGF)38 c3i6pF_
not modelled
5.6
20
PDB header: structural proteinChain: F: PDB Molecule: ethanolamine utilization protein eutm;PDBTitle: ethanolamine utilization microcompartment shell subunit, eutm
39 c3o4aC_
not modelled
5.6
5
PDB header: de novo proteinChain: C: PDB Molecule: de novo designed beta-trefoil architecture with symmetricPDBTitle: crystal structure of symfoil-2: de novo designed beta-trefoil2 architecture with symmetric primary structure
40 c3ngkA_
not modelled
5.5
25
PDB header: unknown functionChain: A: PDB Molecule: propanediol utilization protein pdua;PDBTitle: pdua from salmonella enterica typhimurium
41 d1r89a3
not modelled
5.3
12
Fold: Ferredoxin-likeSuperfamily: PAP/Archaeal CCA-adding enzyme, C-terminal domainFamily: Archaeal tRNA CCA-adding enzyme42 d2ewha1
not modelled
5.2
20
Fold: Ferredoxin-likeSuperfamily: CcmK-likeFamily: CcmK-like43 d1rk8a_
not modelled
5.2
14
Fold: Ferredoxin-likeSuperfamily: RNA-binding domain, RBDFamily: Canonical RBD