| 1 |
|
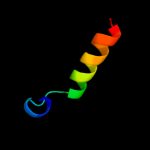
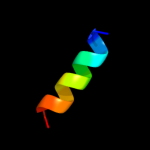
PDB 3sak chain A
Region: 49 - 63
Aligned: 15
Modelled: 15
Confidence: 24.7%
Identity: 40%
Fold: p53 tetramerization domain
Superfamily: p53 tetramerization domain
Family: p53 tetramerization domain
Phyre2
| 2 |
|
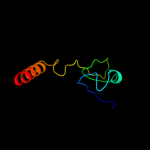
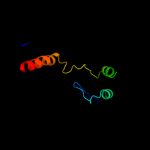
PDB 2j10 chain B
Region: 50 - 63
Aligned: 14
Modelled: 14
Confidence: 23.0%
Identity: 43%
PDB header:transcription
Chain: B: PDB Molecule:cellular tumor antigen p53;
PDBTitle: p53 tetramerization domain mutant t329f q331k
Phyre2
| 3 |
|
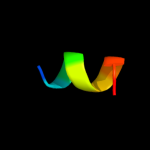
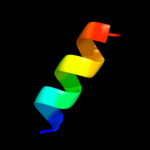
PDB 2j10 chain D
Region: 50 - 63
Aligned: 14
Modelled: 14
Confidence: 23.0%
Identity: 43%
PDB header:transcription
Chain: D: PDB Molecule:cellular tumor antigen p53;
PDBTitle: p53 tetramerization domain mutant t329f q331k
Phyre2
| 4 |
|
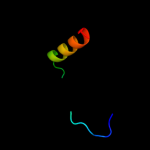
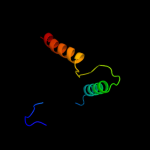
PDB 2j10 chain A
Region: 50 - 63
Aligned: 14
Modelled: 14
Confidence: 23.0%
Identity: 43%
PDB header:transcription
Chain: A: PDB Molecule:cellular tumor antigen p53;
PDBTitle: p53 tetramerization domain mutant t329f q331k
Phyre2
| 5 |
|
PDB 1aie chain A
Region: 50 - 69
Aligned: 20
Modelled: 20
Confidence: 17.1%
Identity: 30%
Fold: p53 tetramerization domain
Superfamily: p53 tetramerization domain
Family: p53 tetramerization domain
Phyre2
| 6 |
|
PDB 2j11 chain D
Region: 50 - 69
Aligned: 20
Modelled: 20
Confidence: 15.5%
Identity: 30%
PDB header:transcription
Chain: D: PDB Molecule:cellular tumor antigen p53;
PDBTitle: p53 tetramerization domain mutant y327s t329g q331g
Phyre2
| 7 |
|
PDB 2k53 chain A
Region: 11 - 33
Aligned: 23
Modelled: 23
Confidence: 11.0%
Identity: 30%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:a3dk08 protein;
PDBTitle: nmr solution structure of a3dk08 protein from clostridium2 thermocellum: northeast structural genomics consortium3 target cmr9
Phyre2
| 8 |
|
PDB 2k5e chain A
Region: 9 - 35
Aligned: 27
Modelled: 27
Confidence: 10.2%
Identity: 15%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: solution structure of putative uncharacterized protein2 gsu1278 from methanocaldococcus jannaschii, northeast3 structural genomics consortium (nesg) target gsr195
Phyre2
| 9 |
|
PDB 1qp8 chain A
Region: 1 - 68
Aligned: 67
Modelled: 67
Confidence: 10.1%
Identity: 15%
PDB header:oxidoreductase
Chain: A: PDB Molecule:formate dehydrogenase;
PDBTitle: crystal structure of a putative formate dehydrogenase from2 pyrobaculum aerophilum
Phyre2
| 10 |
|
PDB 2vzd chain D
Region: 18 - 27
Aligned: 10
Modelled: 10
Confidence: 8.3%
Identity: 60%
PDB header:cell adhesion
Chain: D: PDB Molecule:paxillin;
PDBTitle: crystal structure of the c-terminal calponin homology2 domain of alpha parvin in complex with paxillin ld1 motif
Phyre2
| 11 |
|
PDB 2vzd chain C
Region: 18 - 27
Aligned: 10
Modelled: 10
Confidence: 8.2%
Identity: 60%
PDB header:cell adhesion
Chain: C: PDB Molecule:paxillin;
PDBTitle: crystal structure of the c-terminal calponin homology2 domain of alpha parvin in complex with paxillin ld1 motif
Phyre2
| 12 |
|
PDB 2o8r chain A domain 3
Region: 2 - 26
Aligned: 25
Modelled: 25
Confidence: 7.1%
Identity: 40%
Fold: Phospholipase D/nuclease
Superfamily: Phospholipase D/nuclease
Family: Polyphosphate kinase C-terminal domain
Phyre2
| 13 |
|
PDB 1a1u chain A
Region: 50 - 63
Aligned: 14
Modelled: 14
Confidence: 6.9%
Identity: 36%
Fold: p53 tetramerization domain
Superfamily: p53 tetramerization domain
Family: p53 tetramerization domain
Phyre2
| 14 |
|
PDB 1dxy chain A
Region: 1 - 68
Aligned: 65
Modelled: 68
Confidence: 6.3%
Identity: 20%
PDB header:oxidoreductase
Chain: A: PDB Molecule:d-2-hydroxyisocaproate dehydrogenase;
PDBTitle: structure of d-2-hydroxyisocaproate dehydrogenase
Phyre2
| 15 |
|
PDB 1a6d chain A domain 3
Region: 21 - 33
Aligned: 13
Modelled: 13
Confidence: 6.3%
Identity: 31%
Fold: GroEL-intermediate domain like
Superfamily: GroEL-intermediate domain like
Family: Group II chaperonin (CCT, TRIC), intermediate domain
Phyre2
| 16 |
|
PDB 2o8r chain A
Region: 2 - 60
Aligned: 59
Modelled: 59
Confidence: 6.2%
Identity: 22%
PDB header:transferase
Chain: A: PDB Molecule:polyphosphate kinase;
PDBTitle: crystal structure of polyphosphate kinase from2 porphyromonas gingivalis
Phyre2
| 17 |
|
PDB 2gno chain A domain 1
Region: 51 - 61
Aligned: 11
Modelled: 11
Confidence: 5.5%
Identity: 45%
Fold: post-AAA+ oligomerization domain-like
Superfamily: post-AAA+ oligomerization domain-like
Family: DNA polymerase III clamp loader subunits, C-terminal domain
Phyre2