| 1 |
|
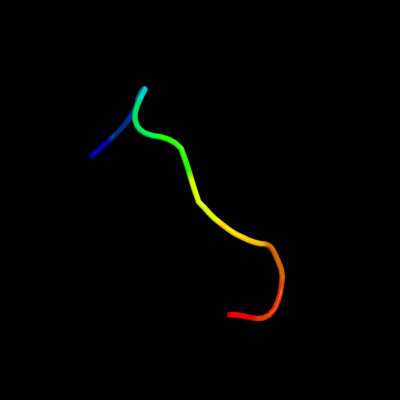
PDB 1c4e chain A
Region: 58 - 67
Aligned: 10
Modelled: 10
Confidence: 36.9%
Identity: 60%
Fold: Knottins (small inhibitors, toxins, lectins)
Superfamily: Gurmarin-like
Family: Gurmarin, a sweet taste-suppressing polypeptide
Phyre2
| 2 |
|
PDB 1a0c chain A
Region: 61 - 71
Aligned: 11
Modelled: 11
Confidence: 20.7%
Identity: 27%
Fold: TIM beta/alpha-barrel
Superfamily: Xylose isomerase-like
Family: Xylose isomerase
Phyre2
| 3 |
|
PDB 1a0e chain A
Region: 61 - 71
Aligned: 11
Modelled: 11
Confidence: 19.0%
Identity: 27%
Fold: TIM beta/alpha-barrel
Superfamily: Xylose isomerase-like
Family: Xylose isomerase
Phyre2
| 4 |
|
PDB 1a0d chain A
Region: 61 - 71
Aligned: 11
Modelled: 11
Confidence: 10.0%
Identity: 27%
Fold: TIM beta/alpha-barrel
Superfamily: Xylose isomerase-like
Family: Xylose isomerase
Phyre2
| 5 |
|
PDB 2vsc chain D
Region: 22 - 71
Aligned: 50
Modelled: 50
Confidence: 9.4%
Identity: 10%
PDB header:cell adhesion
Chain: D: PDB Molecule:leukocyte surface antigen cd47;
PDBTitle: structure of the immunoglobulin-superfamily ectodomain of2 human cd47
Phyre2
| 6 |
|
PDB 1an8 chain A domain 1
Region: 56 - 66
Aligned: 11
Modelled: 11
Confidence: 8.4%
Identity: 36%
Fold: OB-fold
Superfamily: Bacterial enterotoxins
Family: Superantigen toxins, N-terminal domain
Phyre2
| 7 |
|
PDB 1ty0 chain A domain 1
Region: 56 - 66
Aligned: 11
Modelled: 11
Confidence: 7.4%
Identity: 36%
Fold: OB-fold
Superfamily: Bacterial enterotoxins
Family: Superantigen toxins, N-terminal domain
Phyre2
| 8 |
|
PDB 2c2p chain A
Region: 21 - 66
Aligned: 33
Modelled: 33
Confidence: 7.3%
Identity: 27%
PDB header:hydrolase
Chain: A: PDB Molecule:g/u mismatch-specific dna glycosylase;
PDBTitle: the crystal structure of mismatch specific uracil-dna2 glycosylase (mug) from deinococcus radiodurans
Phyre2
| 9 |
|
PDB 2jna chain A domain 1
Region: 1 - 67
Aligned: 67
Modelled: 67
Confidence: 7.0%
Identity: 21%
Fold: Dodecin subunit-like
Superfamily: YdgH-like
Family: YdgH-like
Phyre2
| 10 |
|
PDB 1ujp chain A
Region: 61 - 67
Aligned: 7
Modelled: 7
Confidence: 6.6%
Identity: 43%
Fold: TIM beta/alpha-barrel
Superfamily: Ribulose-phoshate binding barrel
Family: Tryptophan biosynthesis enzymes
Phyre2
| 11 |
|
PDB 1l0n chain K
Region: 64 - 68
Aligned: 5
Modelled: 5
Confidence: 5.9%
Identity: 60%
Fold: Single transmembrane helix
Superfamily: Subunit XI (6.4 kDa protein) of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)
Family: Subunit XI (6.4 kDa protein) of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)
Phyre2
| 12 |
|
PDB 2fyu chain K domain 1
Region: 64 - 68
Aligned: 5
Modelled: 5
Confidence: 5.8%
Identity: 80%
Fold: Single transmembrane helix
Superfamily: Subunit XI (6.4 kDa protein) of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)
Family: Subunit XI (6.4 kDa protein) of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)
Phyre2
| 13 |
|
PDB 3r24 chain A
Region: 32 - 58
Aligned: 22
Modelled: 27
Confidence: 5.8%
Identity: 36%
PDB header:transferase, viral protein
Chain: A: PDB Molecule:2'-o-methyl transferase;
PDBTitle: crystal structure of nsp10/nsp16 complex of sars coronavirus" if2 possible
Phyre2
| 14 |
|
PDB 1eg3 chain A domain 3
Region: 64 - 70
Aligned: 7
Modelled: 7
Confidence: 5.7%
Identity: 71%
Fold: WW domain-like
Superfamily: WW domain
Family: WW domain
Phyre2