| 1 |
|
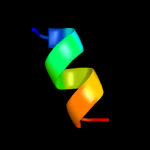
PDB 1wq6 chain A
Region: 36 - 72
Aligned: 28
Modelled: 37
Confidence: 42.0%
Identity: 29%
PDB header:oncoprotein
Chain: A: PDB Molecule:aml1-eto;
PDBTitle: the tetramer structure of the nervy homolgy two (nhr2) domain of aml1-2 eto is critical for aml1-eto's activity
Phyre2
| 2 |
|
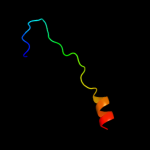
PDB 2kk1 chain A
Region: 24 - 70
Aligned: 39
Modelled: 47
Confidence: 38.4%
Identity: 31%
PDB header:transferase
Chain: A: PDB Molecule:tyrosine-protein kinase abl2;
PDBTitle: solution structure of c-terminal domain of tyrosine-protein2 kinase abl2 from homo sapiens, northeast structural3 genomics consortium (nesg) target hr5537a
Phyre2
| 3 |
|
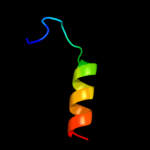
PDB 1zzp chain A
Region: 24 - 70
Aligned: 39
Modelled: 47
Confidence: 26.1%
Identity: 31%
PDB header:transferase
Chain: A: PDB Molecule:proto-oncogene tyrosine-protein kinase abl1;
PDBTitle: solution structure of the f-actin binding domain of bcr-2 abl/c-abl
Phyre2
| 4 |
|
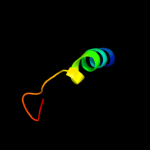
PDB 1a11 chain A
Region: 58 - 67
Aligned: 10
Modelled: 10
Confidence: 23.1%
Identity: 70%
PDB header:acetylcholine receptor
Chain: A: PDB Molecule:acetylcholine receptor m2;
PDBTitle: nmr structure of membrane spanning segment 2 of the2 acetylcholine receptor in dpc micelles, 10 structures
Phyre2
| 5 |
|
PDB 1eq8 chain C
Region: 58 - 67
Aligned: 10
Modelled: 10
Confidence: 18.8%
Identity: 70%
PDB header:signaling protein
Chain: C: PDB Molecule:acetylcholine receptor protein;
PDBTitle: three-dimensional structure of the pentameric helical2 bundle of the acetylcholine receptor m2 transmembrane3 segment
Phyre2
| 6 |
|
PDB 1eq8 chain E
Region: 58 - 67
Aligned: 10
Modelled: 10
Confidence: 18.8%
Identity: 70%
PDB header:signaling protein
Chain: E: PDB Molecule:acetylcholine receptor protein;
PDBTitle: three-dimensional structure of the pentameric helical2 bundle of the acetylcholine receptor m2 transmembrane3 segment
Phyre2
| 7 |
|
PDB 1eq8 chain A
Region: 58 - 67
Aligned: 10
Modelled: 10
Confidence: 18.8%
Identity: 70%
PDB header:signaling protein
Chain: A: PDB Molecule:acetylcholine receptor protein;
PDBTitle: three-dimensional structure of the pentameric helical2 bundle of the acetylcholine receptor m2 transmembrane3 segment
Phyre2
| 8 |
|
PDB 1eq8 chain D
Region: 58 - 67
Aligned: 10
Modelled: 10
Confidence: 18.8%
Identity: 70%
PDB header:signaling protein
Chain: D: PDB Molecule:acetylcholine receptor protein;
PDBTitle: three-dimensional structure of the pentameric helical2 bundle of the acetylcholine receptor m2 transmembrane3 segment
Phyre2
| 9 |
|
PDB 1eq8 chain B
Region: 58 - 67
Aligned: 10
Modelled: 10
Confidence: 18.8%
Identity: 70%
PDB header:signaling protein
Chain: B: PDB Molecule:acetylcholine receptor protein;
PDBTitle: three-dimensional structure of the pentameric helical2 bundle of the acetylcholine receptor m2 transmembrane3 segment
Phyre2
| 10 |
|
PDB 1m5q chain 1
Region: 19 - 45
Aligned: 27
Modelled: 27
Confidence: 10.5%
Identity: 22%
Fold: Sm-like fold
Superfamily: Sm-like ribonucleoproteins
Family: Sm motif of small nuclear ribonucleoproteins, SNRNP
Phyre2
| 11 |
|
PDB 2fna chain A domain 1
Region: 29 - 49
Aligned: 21
Modelled: 21
Confidence: 8.7%
Identity: 19%
Fold: DNA/RNA-binding 3-helical bundle
Superfamily: "Winged helix" DNA-binding domain
Family: Helicase DNA-binding domain
Phyre2
| 12 |
|
PDB 3e0z chain B
Region: 35 - 58
Aligned: 24
Modelled: 23
Confidence: 8.4%
Identity: 29%
PDB header:unknown function
Chain: B: PDB Molecule:protein of unknown function;
PDBTitle: crystal structure of a putative imidazole glycerol phosphate synthase2 homolog (eubrec_1070) from eubacterium rectale at 1.75 a resolution
Phyre2
| 13 |
|
PDB 1izl chain J
Region: 31 - 37
Aligned: 7
Modelled: 7
Confidence: 5.7%
Identity: 57%
PDB header:photosynthesis
Chain: J: PDB Molecule:photosystem ii: subunit psba;
PDBTitle: crystal structure of photosystem ii
Phyre2
| 14 |
|
PDB 1dfw chain A
Region: 38 - 50
Aligned: 13
Modelled: 13
Confidence: 5.6%
Identity: 38%
PDB header:immune system
Chain: A: PDB Molecule:lung surfactant protein b;
PDBTitle: conformational mapping of the n-terminal segment of2 surfactant protein b in lipid using 13c-enhanced fourier3 transform infrared spectroscopy (ftir)
Phyre2
| 15 |
|
PDB 1mu2 chain A
Region: 12 - 50
Aligned: 38
Modelled: 39
Confidence: 5.3%
Identity: 16%
PDB header:transferase
Chain: A: PDB Molecule:hiv-2 rt;
PDBTitle: crystal structure of hiv-2 reverse transcriptase
Phyre2
| 16 |
|
PDB 3blh chain B domain 1
Region: 38 - 69
Aligned: 26
Modelled: 32
Confidence: 5.2%
Identity: 15%
Fold: Cyclin-like
Superfamily: Cyclin-like
Family: Cyclin
Phyre2
| 17 |
|
PDB 3ctd chain B
Region: 14 - 29
Aligned: 16
Modelled: 16
Confidence: 5.1%
Identity: 31%
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:putative atpase, aaa family;
PDBTitle: crystal structure of a putative aaa family atpase from2 prochlorococcus marinus subsp. pastoris
Phyre2