| 1 |
|
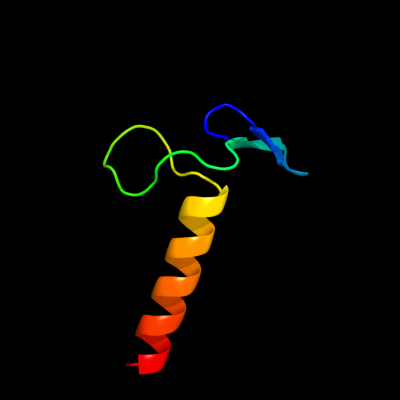
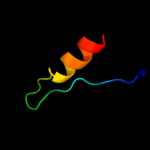
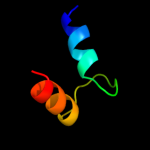
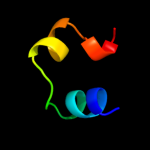
PDB 2ex3 chain B domain 1
Region: 1 - 46
Aligned: 36
Modelled: 46
Confidence: 41.8%
Identity: 22%
Fold: DNA terminal protein
Superfamily: DNA terminal protein
Family: DNA terminal protein
Phyre2
| 2 |
|
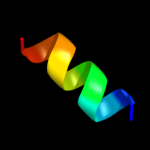
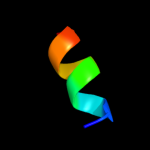
PDB 2kbi chain A
Region: 88 - 116
Aligned: 29
Modelled: 29
Confidence: 34.7%
Identity: 24%
PDB header:metal binding protein
Chain: A: PDB Molecule:sodium channel protein type 5 subunit alpha;
PDBTitle: solution nmr structure of the c-terminal ef-hand domain of2 human cardiac sodium channel nav1.5
Phyre2
| 3 |
|
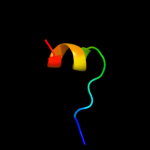
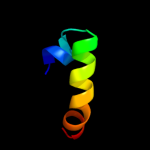
PDB 1wi0 chain A
Region: 40 - 93
Aligned: 44
Modelled: 54
Confidence: 22.9%
Identity: 23%
Fold: beta-Grasp (ubiquitin-like)
Superfamily: CAD & PB1 domains
Family: PB1 domain
Phyre2
| 4 |
|
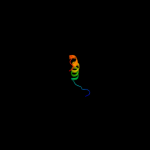
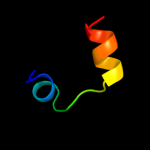
PDB 2npt chain A domain 1
Region: 68 - 93
Aligned: 20
Modelled: 26
Confidence: 14.9%
Identity: 40%
Fold: beta-Grasp (ubiquitin-like)
Superfamily: CAD & PB1 domains
Family: PB1 domain
Phyre2
| 5 |
|
PDB 1m98 chain A
Region: 20 - 31
Aligned: 12
Modelled: 12
Confidence: 13.3%
Identity: 42%
PDB header:unknown function
Chain: A: PDB Molecule:orange carotenoid protein;
PDBTitle: crystal structure of orange carotenoid protein
Phyre2
| 6 |
|
PDB 2e1n chain A
Region: 102 - 117
Aligned: 16
Modelled: 16
Confidence: 13.0%
Identity: 0%
PDB header:circadian clock protein
Chain: A: PDB Molecule:pex;
PDBTitle: crystal structure of the cyanobacterium circadian clock modifier pex
Phyre2
| 7 |
|
PDB 3m6j chain D
Region: 21 - 32
Aligned: 12
Modelled: 12
Confidence: 12.7%
Identity: 42%
PDB header:structural genomics, unknown function
Chain: D: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of unknown function protein from leptospirillum2 rubarum
Phyre2
| 8 |
|
PDB 1nwd chain B
Region: 107 - 120
Aligned: 14
Modelled: 14
Confidence: 11.1%
Identity: 36%
PDB header:binding protein/hydrolase
Chain: B: PDB Molecule:glutamate decarboxylase;
PDBTitle: solution structure of ca2+/calmodulin bound to the c-2 terminal domain of petunia glutamate decarboxylase
Phyre2
| 9 |
|
PDB 1nwd chain C
Region: 107 - 120
Aligned: 14
Modelled: 14
Confidence: 11.1%
Identity: 36%
PDB header:binding protein/hydrolase
Chain: C: PDB Molecule:glutamate decarboxylase;
PDBTitle: solution structure of ca2+/calmodulin bound to the c-2 terminal domain of petunia glutamate decarboxylase
Phyre2
| 10 |
|
PDB 2kbc chain B
Region: 96 - 110
Aligned: 15
Modelled: 15
Confidence: 10.6%
Identity: 47%
PDB header:hormone
Chain: B: PDB Molecule:insl5_b-chain;
PDBTitle: solution structure of human insulin-like peptide 5 (insl5)
Phyre2
| 11 |
|
PDB 2cy5 chain A domain 1
Region: 42 - 102
Aligned: 54
Modelled: 61
Confidence: 9.5%
Identity: 20%
Fold: PH domain-like barrel
Superfamily: PH domain-like
Family: Phosphotyrosine-binding domain (PTB)
Phyre2
| 12 |
|
PDB 3obk chain H
Region: 12 - 38
Aligned: 26
Modelled: 27
Confidence: 9.1%
Identity: 8%
PDB header:lyase
Chain: H: PDB Molecule:delta-aminolevulinic acid dehydratase;
PDBTitle: crystal structure of delta-aminolevulinic acid dehydratase2 (porphobilinogen synthase) from toxoplasma gondii me49 in complex3 with the reaction product porphobilinogen
Phyre2
| 13 |
|
PDB 2dql chain A
Region: 102 - 118
Aligned: 17
Modelled: 17
Confidence: 8.6%
Identity: 6%
PDB header:circadian clock protein
Chain: A: PDB Molecule:pex protein;
PDBTitle: crytal structure of the circadian clock associated protein2 pex from anabaena
Phyre2
| 14 |
|
PDB 1k9u chain A
Region: 84 - 113
Aligned: 30
Modelled: 30
Confidence: 8.1%
Identity: 13%
Fold: EF Hand-like
Superfamily: EF-hand
Family: Polcalcin
Phyre2
| 15 |
|
PDB 1rij chain A
Region: 21 - 30
Aligned: 10
Modelled: 10
Confidence: 6.6%
Identity: 30%
PDB header:de novo protein
Chain: A: PDB Molecule:e6apn1 peptide;
PDBTitle: e6-bind trp-cage (e6apn1)
Phyre2
| 16 |
|
PDB 3c4r chain C
Region: 23 - 49
Aligned: 26
Modelled: 27
Confidence: 5.6%
Identity: 8%
PDB header:structural genomics, unknown function
Chain: C: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of an uncharacterized protein encoded by2 cryptic prophage
Phyre2
| 17 |
|
PDB 2qv6 chain D
Region: 25 - 45
Aligned: 20
Modelled: 21
Confidence: 5.5%
Identity: 15%
PDB header:hydrolase
Chain: D: PDB Molecule:gtp cyclohydrolase iii;
PDBTitle: gtp cyclohydrolase iii from m. jannaschii (mj0145)2 complexed with gtp and metal ions
Phyre2
| 18 |
|
PDB 1tp6 chain A
Region: 21 - 44
Aligned: 22
Modelled: 24
Confidence: 5.5%
Identity: 23%
Fold: Cystatin-like
Superfamily: NTF2-like
Family: PA1314-like
Phyre2