| 1 |
|
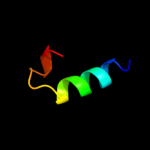
PDB 1o98 chain A domain 1
Region: 7 - 91
Aligned: 71
Modelled: 71
Confidence: 17.9%
Identity: 18%
Fold: 2,3-Bisphosphoglycerate-independent phosphoglycerate mutase, substrate-binding domain
Superfamily: 2,3-Bisphosphoglycerate-independent phosphoglycerate mutase, substrate-binding domain
Family: 2,3-Bisphosphoglycerate-independent phosphoglycerate mutase, substrate-binding domain
Phyre2
| 2 |
|
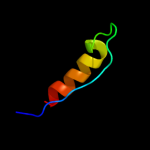
PDB 2k5e chain A
Region: 60 - 86
Aligned: 27
Modelled: 27
Confidence: 17.8%
Identity: 15%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: solution structure of putative uncharacterized protein2 gsu1278 from methanocaldococcus jannaschii, northeast3 structural genomics consortium (nesg) target gsr195
Phyre2
| 3 |
|
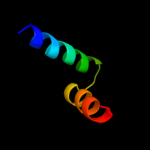
PDB 2qnf chain B
Region: 65 - 92
Aligned: 28
Modelled: 28
Confidence: 17.1%
Identity: 18%
PDB header:hydrolase/dna
Chain: B: PDB Molecule:recombination endonuclease vii;
PDBTitle: crystal structure of t4 endonuclease vii h43n mutant in2 complex with heteroduplex dna containing base mismatches
Phyre2
| 4 |
|
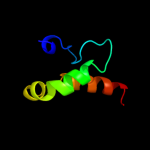
PDB 2bby chain A
Region: 72 - 89
Aligned: 18
Modelled: 18
Confidence: 16.7%
Identity: 11%
Fold: DNA/RNA-binding 3-helical bundle
Superfamily: "Winged helix" DNA-binding domain
Family: DNA-binding domain from rap30
Phyre2
| 5 |
|
PDB 2iqc chain A
Region: 65 - 85
Aligned: 21
Modelled: 21
Confidence: 15.0%
Identity: 24%
PDB header:protein binding
Chain: A: PDB Molecule:fanconi anemia group f protein;
PDBTitle: crystal structure of human fancf protein that functions in2 the assembly of a dna damage signaling complex
Phyre2
| 6 |
|
PDB 2k53 chain A
Region: 60 - 85
Aligned: 26
Modelled: 26
Confidence: 13.6%
Identity: 15%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:a3dk08 protein;
PDBTitle: nmr solution structure of a3dk08 protein from clostridium2 thermocellum: northeast structural genomics consortium3 target cmr9
Phyre2
| 7 |
|
PDB 2ebf chain X domain 2
Region: 58 - 70
Aligned: 13
Modelled: 13
Confidence: 10.5%
Identity: 23%
Fold: EreA/ChaN-like
Superfamily: EreA/ChaN-like
Family: PMT domain-like
Phyre2
| 8 |
|
PDB 5mdh chain A domain 1
Region: 71 - 91
Aligned: 21
Modelled: 21
Confidence: 10.2%
Identity: 5%
Fold: NAD(P)-binding Rossmann-fold domains
Superfamily: NAD(P)-binding Rossmann-fold domains
Family: LDH N-terminal domain-like
Phyre2
| 9 |
|
PDB 2khz chain B
Region: 39 - 69
Aligned: 31
Modelled: 31
Confidence: 8.6%
Identity: 19%
PDB header:nuclear protein
Chain: B: PDB Molecule:c-myc-responsive protein rcl;
PDBTitle: solution structure of rcl
Phyre2
| 10 |
|
PDB 3crc chain B
Region: 52 - 83
Aligned: 32
Modelled: 32
Confidence: 7.6%
Identity: 19%
PDB header:hydrolase
Chain: B: PDB Molecule:protein mazg;
PDBTitle: crystal structure of escherichia coli mazg, the regulator2 of nutritional stress response
Phyre2
| 11 |
|
PDB 3s9v chain D
Region: 34 - 85
Aligned: 44
Modelled: 52
Confidence: 6.5%
Identity: 18%
PDB header:lyase, isomerase
Chain: D: PDB Molecule:abietadiene synthase, chloroplastic;
PDBTitle: abietadiene synthase from abies grandis
Phyre2
| 12 |
|
PDB 2auh chain B
Region: 5 - 22
Aligned: 18
Modelled: 18
Confidence: 5.8%
Identity: 39%
PDB header:transferase/signaling protein
Chain: B: PDB Molecule:growth factor receptor-bound protein 14;
PDBTitle: crystal structure of the grb14 bps region in complex with2 the insulin receptor tyrosine kinase
Phyre2
| 13 |
|
PDB 2vy2 chain A
Region: 47 - 77
Aligned: 31
Modelled: 31
Confidence: 5.7%
Identity: 13%
PDB header:transcription
Chain: A: PDB Molecule:protein leafy;
PDBTitle: structure of leafy transcription factor from arabidopsis2 thaliana in complex with dna from ag-i promoter
Phyre2