| 1 |
|
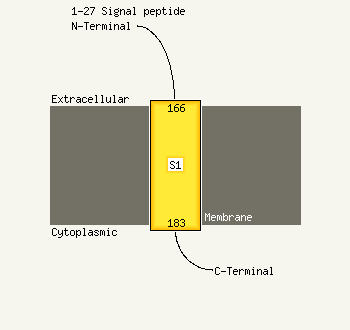
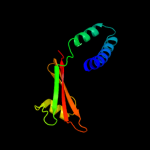
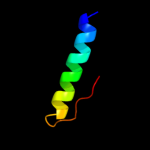
PDB 2asx chain A domain 1
Region: 182 - 213
Aligned: 32
Modelled: 32
Confidence: 97.8%
Identity: 3%
Fold: HAMP domain-like
Superfamily: HAMP domain-like
Family: HAMP domain
Phyre2
| 2 |
|
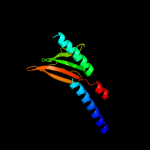
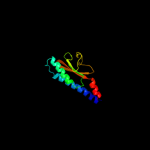
PDB 3by8 chain A domain 1
Region: 39 - 156
Aligned: 118
Modelled: 118
Confidence: 97.3%
Identity: 16%
Fold: Profilin-like
Superfamily: Sensory domain-like
Family: Sensory domain of two-component sensor kinase
Phyre2
| 3 |
|
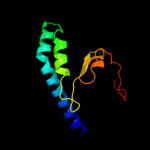
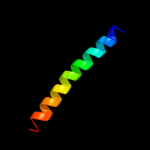
PDB 1p0z chain A
Region: 43 - 152
Aligned: 110
Modelled: 110
Confidence: 97.2%
Identity: 18%
Fold: Profilin-like
Superfamily: Sensory domain-like
Family: Sensory domain of two-component sensor kinase
Phyre2
| 4 |
|
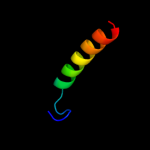
PDB 3by9 chain A
Region: 32 - 153
Aligned: 122
Modelled: 122
Confidence: 54.1%
Identity: 16%
PDB header:transferase
Chain: A: PDB Molecule:sensor protein;
PDBTitle: crystal structure of the v. cholerae histidine kinase dctb2 sensor domain
Phyre2
| 5 |
|
PDB 3pjv chain D
Region: 35 - 157
Aligned: 119
Modelled: 123
Confidence: 48.0%
Identity: 13%
PDB header:lyase
Chain: D: PDB Molecule:cyclic dimeric gmp binding protein;
PDBTitle: structure of pseudomonas fluorescence lapd periplasmic domain
Phyre2
| 6 |
|
PDB 2p7j chain A domain 2
Region: 33 - 155
Aligned: 123
Modelled: 123
Confidence: 43.0%
Identity: 15%
Fold: Profilin-like
Superfamily: Sensory domain-like
Family: YkuI C-terminal domain-like
Phyre2
| 7 |
|
PDB 3b42 chain B
Region: 42 - 152
Aligned: 109
Modelled: 111
Confidence: 31.7%
Identity: 18%
PDB header:signaling protein
Chain: B: PDB Molecule:methyl-accepting chemotaxis protein, putative;
PDBTitle: periplasmic sensor domain of chemotaxis protein gsu0935
Phyre2
| 8 |
|
PDB 3b47 chain A
Region: 43 - 149
Aligned: 104
Modelled: 107
Confidence: 28.8%
Identity: 19%
PDB header:signaling protein
Chain: A: PDB Molecule:methyl-accepting chemotaxis protein;
PDBTitle: periplasmic sensor domain of chemotaxis protein gsu0582
Phyre2
| 9 |
|
PDB 3e4p chain B
Region: 41 - 156
Aligned: 116
Modelled: 116
Confidence: 25.8%
Identity: 19%
PDB header:transferase
Chain: B: PDB Molecule:c4-dicarboxylate transport sensor protein dctb;
PDBTitle: crystal structure of malonate occupied dctb
Phyre2
| 10 |
|
PDB 2qhk chain A
Region: 33 - 123
Aligned: 88
Modelled: 91
Confidence: 25.5%
Identity: 11%
PDB header:signaling protein
Chain: A: PDB Molecule:methyl-accepting chemotaxis protein;
PDBTitle: crystal structure of methyl-accepting chemotaxis protein from vibrio2 parahaemolyticus rimd 2210633
Phyre2
| 11 |
|
PDB 2kse chain A
Region: 3 - 29
Aligned: 27
Modelled: 27
Confidence: 20.3%
Identity: 22%
PDB header:transferase
Chain: A: PDB Molecule:sensor protein qsec;
PDBTitle: backbone structure of the membrane domain of e. coli2 histidine kinase receptor qsec, center for structures of3 membrane proteins (csmp) target 4311c
Phyre2
| 12 |
|
PDB 1u6k chain A domain 1
Region: 180 - 214
Aligned: 35
Modelled: 35
Confidence: 19.7%
Identity: 6%
Fold: F420-dependent methylenetetrahydromethanopterin dehydrogenase (MTD)
Superfamily: F420-dependent methylenetetrahydromethanopterin dehydrogenase (MTD)
Family: F420-dependent methylenetetrahydromethanopterin dehydrogenase (MTD)
Phyre2
| 13 |
|
PDB 3lnr chain A
Region: 179 - 207
Aligned: 29
Modelled: 29
Confidence: 18.0%
Identity: 7%
PDB header:signaling protein
Chain: A: PDB Molecule:aerotaxis transducer aer2;
PDBTitle: crystal structure of poly-hamp domains from the p. aeruginosa soluble2 receptor aer2
Phyre2
| 14 |
|
PDB 3fos chain A
Region: 30 - 158
Aligned: 125
Modelled: 129
Confidence: 17.0%
Identity: 18%
PDB header:transferase
Chain: A: PDB Molecule:sensor protein;
PDBTitle: crystal structure of two-component sensor histidine kinase domain from2 bacillus subtilis subsp. subtilis str. 168
Phyre2
| 15 |
|
PDB 3hd7 chain A
Region: 3 - 33
Aligned: 31
Modelled: 31
Confidence: 10.5%
Identity: 10%
PDB header:exocytosis
Chain: A: PDB Molecule:vesicle-associated membrane protein 2;
PDBTitle: helical extension of the neuronal snare complex into the membrane,2 spacegroup c 1 2 1
Phyre2
| 16 |
|
PDB 1sxj chain A domain 1
Region: 187 - 202
Aligned: 16
Modelled: 16
Confidence: 6.3%
Identity: 0%
Fold: post-AAA+ oligomerization domain-like
Superfamily: post-AAA+ oligomerization domain-like
Family: DNA polymerase III clamp loader subunits, C-terminal domain
Phyre2
| 17 |
|
PDB 1hp9 chain A
Region: 209 - 214
Aligned: 6
Modelled: 6
Confidence: 5.7%
Identity: 33%
PDB header:toxin
Chain: A: PDB Molecule:kappa-hefutoxin 1;
PDBTitle: kappa-hefutoxins: a novel class of potassium channel toxins2 from scorpion venom
Phyre2