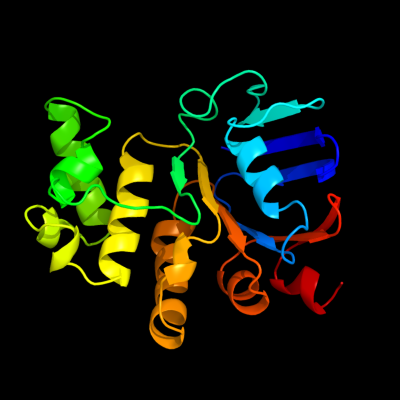
| | |
3 | . | . | . | . | . | . | 10 | . | . | . | . | |
|
|
|
. | . | . | . | . | 20 | . | . | . | . | . | . | . | . | . | 30 | . | . | . | . | . | . | . | . | . | 40 | . | . | . | . | . | . | . | . | . | 50 | |
. | . | . | . | . | . | . | . | . | 60 | . | . | . | . | . | . | . | . | . | 70 | . | . | . | . | . | . | . |
| Predicted Secondary structure | |
 |  |  |  |  |  |
|
|  |  |  |  | . | . | . | . |  |  |  |
|
|  |  |  |  |
|
|
|
|  |  |  |  |  |
|
|
|
|
|
|  |  |  |  |  |  |  |  |  |
|
|
| . |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| Query SS confidence | |
|
|
|
|
|
|
|
|
|
|
|
| . |
. |
. |
. |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| . |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| Query Sequence | |
I | V | M | Q | L | Q | D | V | A | E | S | T | . |
. |
. |
. |
R | L | G | P | L | S | G | E | V | R | A | G | E | I | L | H | L | V | G | P | N | G | A | G | K | S | T | L | L | A | R | M | A | G | M | T | . |
S | G | K | G | S | I | Q | F | A | G | Q | P | L | E | A | W | S | A | T | K | L | A | L | H | R | A | Y |
| Query Conservation | |
| |
| |
| |
|
|
| | | | . | . | . | . | |
| | |
|
|
| |
| | |
|
|
| |
|
|
|
| |
|
|
|
|
|
|
|
|
| | |
| |
|
| | . | | | |
| |
| | | | | | | | | | | | | | | | | | | | | |
| Alig confidence | |
|
|
|
|
|
|
|
|
|
|
|
| . | . | . | . |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| . |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| . | . |
|
|
|
|
|
| Template Conservation | |
| |
| | | | |
| |
| | | | | | | |
| | |
|
|
| |
| | |
|
|
| | |
| |
| |
|
|
|
|
|
|
|
|
| | |
| |
| | | |
| | |
| |
| |
| |
| | | | | | | | | | | . | . | | |
| | |
| Template Sequence | |
V | E | I | K | L | E | N | I | V | K | K | F | G | N | F | T | A | L | N | N | I | N | L | K | I | K | D | G | E | F | M | A | L | L | G | P | S | G | S | G | K | S | T | L | L | Y | T | I | A | G | I | Y | K | P | T | S | G | K | I | Y | F | D | E | K | D | V | T | E | L | P | P | K | D | . |
. |
R | N | V | G | L |
| Template Known Secondary structure | |
|
|  |  |  |  |  |  |  |  |  |  | T | T |  |  |  |  |  |  |  |  |  |  |  |
| T | T |
|  |  |  |  |  |
|
| T | T | S | S |  |  |  |  |  |  |  |  |  | T | S | S |
|
| S |  |  |  |  |  |  | T | T |  |  |
| T | T | S |
|  |  |  | . | . |  | T |  |  |  |
| Template Predicted Secondary structure | |
|  |  |  |  |  |  |  |  |  |  |  |
|
|  |  |  |  |  |
|
|  |  |  |  |
|
|
|
|  |  |  |  |  |
|
|
|
|
|
|  |  |  |  |  |  |  |  |  |
|
|
|
|
|
|
|
|  |  |  |  |
|
|  |  |
|
|
|
|
|  |  |  | . | . |
|  |  |  |  |
| Template SS confidence | |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| | |
2 | . | . | . | . | . | . | . | 10 | . | . | . | . | . | . | . | . | . | 20 | . | . | . | . | . | . | . | . | . | 30 | . | . | . | . | . | . | . | . | . | 40 | . | . | . | . | . | . | . | . | . | 50 | . | . | . | . | . | . | . | . | . | 60 | . | . | . | . | . | . | . | . | . | 70 | . | . | . | . | |
|
. | . | . | . | . |
| |
| | |
78 | . | 80 | . | . | . | . | . | . | . | . | . | 90 | . | . | . | . | . | . | |
|
|
|
|
. | . | . | 100 | . | . | . | . | . | . | . | . | . | 110 | . | . | . | . | . | . | . | . | . | 120 | . | . | . | . | . | . | . | . | . | 130 | . | . | . | . | . | . | . | . | . | 140 | . | . | . | . | . | . | . | . | . | 150 | . | . |
| Predicted Secondary structure | |
|
|
|
|
|
|
|
|
|
|
|
|  |  |  |  |  |  |  | . | . | . | . | . |  |
|
|
|
|  |  |  |  |  |  |  |  |  |  |  |  |
|
|
|
|
|  |  |
|
|
|
|
|
|
|
|
|
|
|  |  |  |  |  |  |  |  |  |  |  |
|
|
|  |  |  |
|
|
|
|
| Query SS confidence | |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| . |
. |
. |
. |
. |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| Query Sequence | |
L | S | Q | Q | Q | T | P | P | F | A | T | P | V | W | H | Y | L | T | L | . |
. |
. |
. |
. |
H | Q | H | D | K | T | R | T | E | L | L | N | D | V | A | G | A | L | A | L | D | D | K | L | G | R | S | T | N | Q | L | S | G | G | E | W | Q | R | V | R | L | A | A | V | V | L | Q | I | T | P | Q | A | N | P | A | G |
| Query Conservation | |
| | | | | | | | | | |
|
| | | |
| | | . | . | . | . | . | | | | | | | | | | | | | | | | | | | | | | | | | | | | | | |
|
| |
| | | | | | | |
| | |
| | | |
| | |
| | | |
|
| Alig confidence | |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| . | . | . | . | . |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| . | . | . | . | . | . | . |
|
|
| Template Conservation | |
| |
| | | |
| | | |
|
|
| | |
| | | | | | | | | | | | | | | | | | | | | | |
| | | | |
| | | | | | | | | | |
|
| |
| | |
|
|
| |
|
|
|
|
| | | . | . | . | . | . | . | . | |
|
| Template Sequence | |
V | F | Q | N | W | A | L | Y | P | H | M | T | V | Y | K | N | I | A | F | P | L | E | L | R | K | A | P | R | E | E | I | D | K | K | V | R | E | V | A | K | M | L | H | I | D | K | L | L | N | R | Y | P | W | Q | L | S | G | G | Q | Q | Q | R | V | A | I | A | R | A | L | V | K | . |
. |
. |
. |
. |
. |
. |
E | P |
| Template Known Secondary structure | |
 |
| T | T |
|
|
|
| T | T | S |
|  |  |  |  |  |  |  |  |  |  |  | T | T |
|
|  |  |  |  |  |  |  |  |  |  |  |  |  |  | T | T |
| G | G | G | T | T | S |
| G | G | G | S |
|  |  |  |  |  |  |  |  |  |  |  |  |  | T | T | . | . | . | . | . | . | . |
|
|
| Template Predicted Secondary structure | |
 |  |  |
|
|
|
|
|
|
|  |  |  |  |  |
|
|
|
|
|
|
|
|
|
|
|
|  |  |  |  |  |  |  |  |  |  |  |  |  |  |
|
|
|
|
|  |  |  |  |
|
|
|
|
|
|
|  |  |  |  |  |  |  |  |  |  |  |  |  |
| . | . | . | . | . | . | . |
|
|
| Template SS confidence | |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| | |
80 | . | . | . | . | . | . | . | . | . | 90 | . | . | . | . | . | . | . | . | . | 100 | . | . | . | . | . | . | . | . | . | 110 | . | . | . | . | . | . | . | . | . | 120 | . | . | . | . | . | . | . | . | . | 130 | . | . | . | . | . | . | . | . | . | 140 | . | . | . | . | . | . | . | . | . | 150 | |
|
|
|
|
|
|
. | . |
| |
| | |
153 | . | . | . | . | . | . | 160 | . | . | . | . | . | . | . | . | . | 170 | . | . | . | . | . | . | . | . | . | |
180 | . | . | . | . | . | . | . | . | . | 190 | . | . | . | . | . | . | . | . | . | 200 | . | . | . | . | . | . | . | . | . | 210 | . | . | . | . | . | . | . | . | . | 220 | . | . | . | . |
| Predicted Secondary structure | |
 |  |  |  |  |  |
|
|
|
|
|
|
|  |  |  |  |  |  |  |  |  |  |  |  |  |  | . |  |  |  |
|
|  |  |  |  |  |  |
|
|  |  |  |  |  |  |  |
|
|  |  |  |  |  |  |
|
|  |  |  |  |  |
|
|  |  |  |  |  |
|
|  |
| Query SS confidence | |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| . |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| Query Sequence | |
Q | L | L | L | L | D | E | P | M | N | S | L | D | V | A | Q | Q | S | A | L | D | K | I | L | S | A | L | . |
C | Q | Q | G | L | A | I | V | M | S | S | H | D | L | N | H | T | L | R | H | A | H | R | A | W | L | L | K | G | G | K | M | L | A | S | G | R | R | E | E | V | L | T | P | P |
| Query Conservation | |
|
|
|
|
|
|
|
|
| |
|
|
| | | | | | |
| | | |
| | |
| . | | | |
| |
|
|
| | |
|
| | | | | | | | | |
|
|
| |
|
| | |
| |
|
| | |
| | | |
|
| | | | |
| Alig confidence | |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| . |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| Template Conservation | |
|
|
|
|
|
|
|
|
|
| |
|
| | | | | | |
| | | |
| | | | | | | |
| |
|
|
| |
|
|
| | | |
| | | | | |
| |
| |
|
| | |
| |
| | | |
| | | |
|
| | | |
|
| Template Sequence | |
E | V | L | L | L | D | E | P | L | S | N | L | D | A | L | L | R | L | E | V | R | A | E | L | K | R | L | Q | K | E | L | G | I | T | T | V | Y | V | T | H | D | Q | A | E | A | L | A | M | A | D | R | I | A | V | I | R | E | G | E | I | L | Q | V | G | T | P | D | E | V | Y | Y | K | P |
| Template Known Secondary structure | |
S |  |  |  |  |  | S | G | G | G | G | S |
|  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  | T |
|  |  |  |  |  |  | S |
|  |  |  |  |  |  |  |
| S |  |  |  |  |  |  | T | T |  |  |  |  |  |  |
|  |  |  |  |  |  | S |
|
| Template Predicted Secondary structure | |
 |  |  |  |  |  |
|
|
|
|
|
|
|  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |
|
|
|
|  |  |  |  |  |  |
|  |  |  |  |  |  |  |
|
|  |  |  |  |
|
|
|
|  |  |  |  |  |
|
|  |  |  |  |  |  |
|
|
| Template SS confidence | |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| | |
153 | . | . | . | . | . | . | 160 | . | . | . | . | . | . | . | . | . | 170 | . | . | . | . | . | . | . | . | . | 180 | . | . | . | . | . | . | . | . | . | 190 | . | . | . | . | . | . | . | . | . | 200 | . | . | . | . | . | . | . | . | . | 210 | . | . | . | . | . | . | . | . | . | 220 | . | . | . | . | . |
| |