| 1 |
|
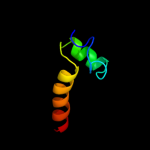
PDB 2w83 chain C
Region: 33 - 45
Aligned: 13
Modelled: 13
Confidence: 53.1%
Identity: 54%
PDB header:protein transport
Chain: C: PDB Molecule:c-jun-amino-terminal kinase-interacting protein
PDBTitle: crystal structure of the arf6 gtpase in complex with a2 specific effector, jip4
Phyre2
| 2 |
|
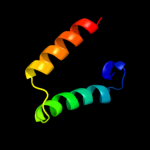
PDB 2xgl chain B
Region: 37 - 47
Aligned: 11
Modelled: 11
Confidence: 9.6%
Identity: 45%
PDB header:antibiotic
Chain: B: PDB Molecule:colicin-m immunity protein;
PDBTitle: the x-ray structure of the escherichia coli colicin m immunity2 protein demonstrates the presence of a disulphide bridge, which is3 functionally essential
Phyre2
| 3 |
|
PDB 1tzz chain A domain 2
Region: 30 - 47
Aligned: 18
Modelled: 18
Confidence: 8.1%
Identity: 17%
Fold: Enolase N-terminal domain-like
Superfamily: Enolase N-terminal domain-like
Family: Enolase N-terminal domain-like
Phyre2
| 4 |
|
PDB 1yrl chain D
Region: 9 - 18
Aligned: 10
Modelled: 10
Confidence: 7.0%
Identity: 20%
PDB header:oxidoreductase
Chain: D: PDB Molecule:ketol-acid reductoisomerase;
PDBTitle: escherichia coli ketol-acid reductoisomerase
Phyre2
| 5 |
|
PDB 1yey chain A domain 2
Region: 30 - 46
Aligned: 17
Modelled: 17
Confidence: 6.3%
Identity: 12%
Fold: Enolase N-terminal domain-like
Superfamily: Enolase N-terminal domain-like
Family: Enolase N-terminal domain-like
Phyre2
| 6 |
|
PDB 2kzt chain A
Region: 1 - 42
Aligned: 42
Modelled: 42
Confidence: 6.0%
Identity: 17%
PDB header:apoptosis
Chain: A: PDB Molecule:programmed cell death protein 4;
PDBTitle: structure of the tandem ma-3 region of pdcd4
Phyre2
| 7 |
|
PDB 2aqe chain A domain 1
Region: 1 - 44
Aligned: 44
Modelled: 44
Confidence: 5.7%
Identity: 16%
Fold: DNA/RNA-binding 3-helical bundle
Superfamily: Homeodomain-like
Family: SWIRM domain
Phyre2
| 8 |
|
PDB 2rg8 chain A
Region: 1 - 42
Aligned: 42
Modelled: 42
Confidence: 5.7%
Identity: 17%
PDB header:apoptosis, translation
Chain: A: PDB Molecule:programmed cell death protein 4;
PDBTitle: crystal structure of programmed for cell death 4 middle ma32 domain
Phyre2