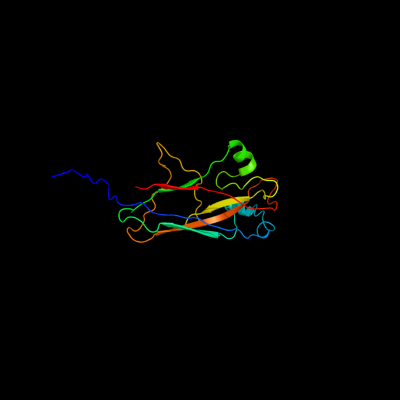
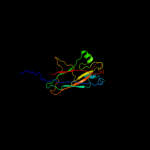
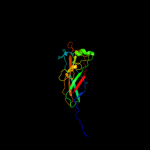
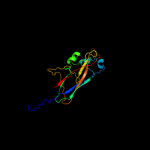
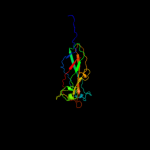
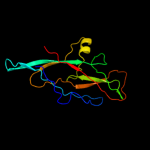
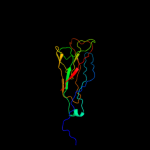
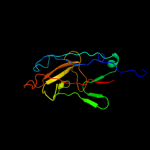
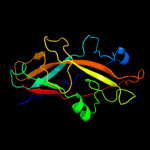
1 c3jwnK_
99.9
16
PDB header: protein binding/cell adhesionChain: K: PDB Molecule: protein fimf;PDBTitle: complex of fimc, fimf, fimg and fimh
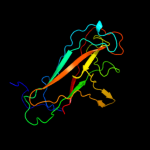
2 c3jwnL_
99.9
16
PDB header: protein binding/cell adhesionChain: L: PDB Molecule: protein fimf;PDBTitle: complex of fimc, fimf, fimg and fimh
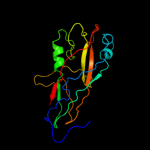
3 c3jwnE_
99.9
16
PDB header: protein binding/cell adhesionChain: E: PDB Molecule: protein fimf;PDBTitle: complex of fimc, fimf, fimg and fimh
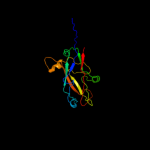
4 c3jwnF_
99.9
16
PDB header: protein binding/cell adhesionChain: F: PDB Molecule: protein fimf;PDBTitle: complex of fimc, fimf, fimg and fimh
5 c2jtyA_
99.9
18
PDB header: structural proteinChain: A: PDB Molecule: type-1 fimbrial protein, a chain;PDBTitle: self-complemented variant of fima, the main subunit of type 1 pilus
6 c2jmrA_
99.9
17
PDB header: cell adhesionChain: A: PDB Molecule: fimf;PDBTitle: nmr structure of the e. coli type 1 pilus subunit fimf
7 d2uy6b1
99.8
11
Fold: Common fold of diphtheria toxin/transcription factors/cytochrome fSuperfamily: Bacterial adhesinsFamily: Pilus subunits8 d2j2zb1
99.8
15
Fold: Common fold of diphtheria toxin/transcription factors/cytochrome fSuperfamily: Bacterial adhesinsFamily: Pilus subunits9 d1pdkb_
99.8
14
Fold: Common fold of diphtheria toxin/transcription factors/cytochrome fSuperfamily: Bacterial adhesinsFamily: Pilus subunits10 c3bwuF_
99.7
16
PDB header: chaperone, structural, membrane proteinChain: F: PDB Molecule: protein fimf;PDBTitle: crystal structure of the ternary complex of fimd (n-terminal domain,2 fimdn) with fimc and the n-terminally truncated pilus subunit fimf3 (fimft)
11 c3bfwA_
99.7
13
PDB header: structural protein/structural proteinChain: A: PDB Molecule: protein fimg;PDBTitle: crystal structure of truncated fimg (fimgt) in complex with the donor2 strand peptide of fimf (dsf)
12 d1ze3h1
99.6
21
Fold: Common fold of diphtheria toxin/transcription factors/cytochrome fSuperfamily: Bacterial adhesinsFamily: Pilus subunits13 c1klfP_
99.5
21
PDB header: chaperone/adhesin complexChain: P: PDB Molecule: fimh protein;PDBTitle: fimh adhesin-fimc chaperone complex with d-mannose
14 c2w07B_
99.5
13
PDB header: cell adhesionChain: B: PDB Molecule: minor pilin subunit papf;PDBTitle: structural determinants of polymerization reactivity of the2 p pilus adaptor subunit papf
15 d1n12a_
99.0
14
Fold: Common fold of diphtheria toxin/transcription factors/cytochrome fSuperfamily: Bacterial adhesinsFamily: Pilus subunits16 d1y0ua_
26.5
25
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: ArsR-like transcriptional regulators17 c1w3gA_
14.5
19
PDB header: toxin/lectinChain: A: PDB Molecule: hemolytic lectin from laetiporus sulphureus;PDBTitle: hemolytic lectin from the mushroom laetiporus sulphureus2 complexed with two n-acetyllactosamine molecules.
18 c3nqnB_
10.7
4
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of a protein with unknown function. (dr_2006) from2 deinococcus radiodurans at 1.88 a resolution
19 c3rpjA_
7.2
58
PDB header: transcription regulatorChain: A: PDB Molecule: curlin genes transcriptional regulator;PDBTitle: structure of a curlin genes transcriptional regulator protein from2 proteus mirabilis hi4320.
20 d3c7ba3
6.5
21
Fold: Nitrite and sulphite reductase 4Fe-4S domain-likeSuperfamily: Nitrite and sulphite reductase 4Fe-4S domain-likeFamily: Nitrite and sulphite reductase 4Fe-4S domain-like21 c1bctA_
not modelled
6.4
21
PDB header: photoreceptorChain: A: PDB Molecule: bacteriorhodopsin;PDBTitle: three-dimensional structure of proteolytic fragment 163-2312 of bacterioopsin determined from nuclear magnetic3 resonance data in solution
22 d2v4ja3
not modelled
6.2
14
Fold: Nitrite and sulphite reductase 4Fe-4S domain-likeSuperfamily: Nitrite and sulphite reductase 4Fe-4S domain-likeFamily: Nitrite and sulphite reductase 4Fe-4S domain-like23 d3c7bb3
not modelled
6.1
16
Fold: Nitrite and sulphite reductase 4Fe-4S domain-likeSuperfamily: Nitrite and sulphite reductase 4Fe-4S domain-likeFamily: Nitrite and sulphite reductase 4Fe-4S domain-like24 d2v4jb3
not modelled
5.8
8
Fold: Nitrite and sulphite reductase 4Fe-4S domain-likeSuperfamily: Nitrite and sulphite reductase 4Fe-4S domain-likeFamily: Nitrite and sulphite reductase 4Fe-4S domain-like25 d1aopa4
not modelled
5.8
12
Fold: Nitrite and sulphite reductase 4Fe-4S domain-likeSuperfamily: Nitrite and sulphite reductase 4Fe-4S domain-likeFamily: Nitrite and sulphite reductase 4Fe-4S domain-like26 d1z1za1
not modelled
5.6
25
Fold: Phage tail protein-likeSuperfamily: Phage tail protein-likeFamily: Lambda phage gpU-like27 d2akja4
not modelled
5.5
12
Fold: Nitrite and sulphite reductase 4Fe-4S domain-likeSuperfamily: Nitrite and sulphite reductase 4Fe-4S domain-likeFamily: Nitrite and sulphite reductase 4Fe-4S domain-like