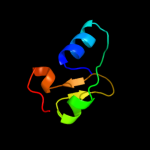
| 1 |
|
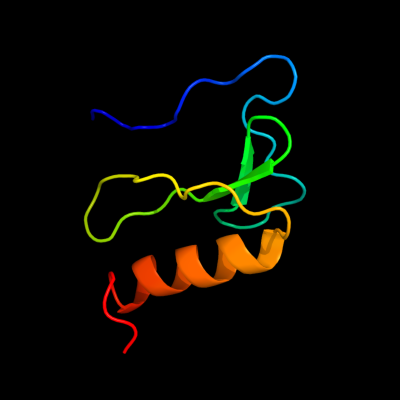
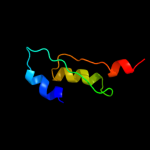
PDB 2bn8 chain A
Region: 14 - 80
Aligned: 67
Modelled: 67
Confidence: 100.0%
Identity: 100%
PDB header:cell cycle protein
Chain: A: PDB Molecule:cell division activator ceda;
PDBTitle: solution structure and interactions of the e.coli cell2 division activator protein ceda
Phyre2
| 2 |
|
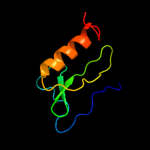
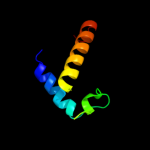
PDB 2d35 chain A
Region: 19 - 80
Aligned: 62
Modelled: 62
Confidence: 100.0%
Identity: 100%
PDB header:cell cycle
Chain: A: PDB Molecule:cell division activator ceda;
PDBTitle: solution structure of cell division reactivation factor,2 ceda
Phyre2
| 3 |
|
PDB 1udx chain A domain 3
Region: 36 - 76
Aligned: 35
Modelled: 41
Confidence: 27.3%
Identity: 29%
Fold: Obg GTP-binding protein C-terminal domain
Superfamily: Obg GTP-binding protein C-terminal domain
Family: Obg GTP-binding protein C-terminal domain
Phyre2
| 4 |
|
PDB 1pii chain A
Region: 7 - 57
Aligned: 48
Modelled: 51
Confidence: 22.4%
Identity: 15%
PDB header:bifunctional(isomerase and synthase)
Chain: A: PDB Molecule:n-(5'phosphoribosyl)anthranilate isomerase;
PDBTitle: three-dimensional structure of the bifunctional enzyme2 phosphoribosylanthranilate isomerase:3 indoleglycerolphosphate synthase from escherichia coli4 refined at 2.0 angstroms resolution
Phyre2
| 5 |
|
PDB 1hh4 chain E
Region: 14 - 45
Aligned: 26
Modelled: 32
Confidence: 11.6%
Identity: 46%
Fold: Immunoglobulin-like beta-sandwich
Superfamily: E set domains
Family: RhoGDI-like
Phyre2
| 6 |
|
PDB 1ds6 chain B
Region: 14 - 45
Aligned: 26
Modelled: 32
Confidence: 8.9%
Identity: 46%
Fold: Immunoglobulin-like beta-sandwich
Superfamily: E set domains
Family: RhoGDI-like
Phyre2
| 7 |
|
PDB 1ajw chain A
Region: 14 - 45
Aligned: 30
Modelled: 32
Confidence: 8.1%
Identity: 33%
Fold: Immunoglobulin-like beta-sandwich
Superfamily: E set domains
Family: RhoGDI-like
Phyre2
| 8 |
|
PDB 1rho chain A
Region: 14 - 45
Aligned: 30
Modelled: 32
Confidence: 7.5%
Identity: 30%
Fold: Immunoglobulin-like beta-sandwich
Superfamily: E set domains
Family: RhoGDI-like
Phyre2
| 9 |
|
PDB 2o2k chain A
Region: 28 - 79
Aligned: 52
Modelled: 52
Confidence: 6.9%
Identity: 27%
PDB header:transferase
Chain: A: PDB Molecule:methionine synthase;
PDBTitle: crystal structure of the activation domain of human2 methionine synthase isoform/mutant d963e/k1071n
Phyre2
| 10 |
|
PDB 1bds chain A
Region: 35 - 41
Aligned: 7
Modelled: 7
Confidence: 6.5%
Identity: 71%
Fold: Defensin-like
Superfamily: Defensin-like
Family: Defensin
Phyre2
| 11 |
|
PDB 1fso chain A
Region: 14 - 45
Aligned: 30
Modelled: 32
Confidence: 5.7%
Identity: 30%
Fold: Immunoglobulin-like beta-sandwich
Superfamily: E set domains
Family: RhoGDI-like
Phyre2
| 12 |
|
PDB 1x60 chain A
Region: 58 - 76
Aligned: 19
Modelled: 19
Confidence: 5.5%
Identity: 32%
PDB header:hydrolase
Chain: A: PDB Molecule:sporulation-specific n-acetylmuramoyl-l-alanine
PDBTitle: solution structure of the peptidoglycan binding domain of2 b. subtilis cell wall lytic enzyme cwlc
Phyre2
| 13 |
|
PDB 1q45 chain A
Region: 29 - 80
Aligned: 43
Modelled: 52
Confidence: 5.3%
Identity: 16%
Fold: TIM beta/alpha-barrel
Superfamily: FMN-linked oxidoreductases
Family: FMN-linked oxidoreductases
Phyre2